Figure 2.
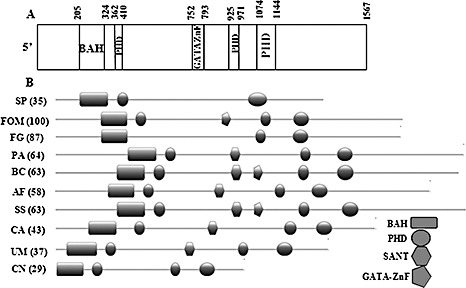
SNT2 protein structure of Fusarium oxysporum f.sp. melonis (FOM). (A) The FOM Snt2 protein carries five conserved domains: a bromo‐adjacent domain (BAH) at amino acid position 205–324; three plant homeodomain Zn fingers (PHD) at positions 362–410, 752–793 and 1074–1144; and a GATA‐type Zn finger at position 925–971. (B) Comparison of the structural domains of SNT2 proteins between different fungi (BAH, rectangle; PHD, circle; GATA Zn finger, pentagon; SANT, hexagon). Sequences shown: SNT2 (YGL131C), Schizosaccharomyces pombe (SP); Snt2, Fusarium oxysporum f.sp. melonis (FOM); FOXG_01993.2, Fusarium oxysporum f.sp. lycopersici (FOL); XP_387009, Fusarium graminearum (FG); XP_001908700, Podospora anserina (PA); XP_001549253, Botrytis cinerea (BC); XP_750273, Aspergillus fumigatus (AF); XP_001584831, Sclerotinia sclerotiorum (SS); XP_716567, Candida albicans (CA); XP_761724, Ustilago maydis (UM); AAN75722 (ZNF1), Cryptococcus neoformans (CN). Similarity (percentage, in parentheses) between SNT2 of FOM and other fungi was analysed by the MatGAT program (Campanella et al., 2003).
