Figure 5.
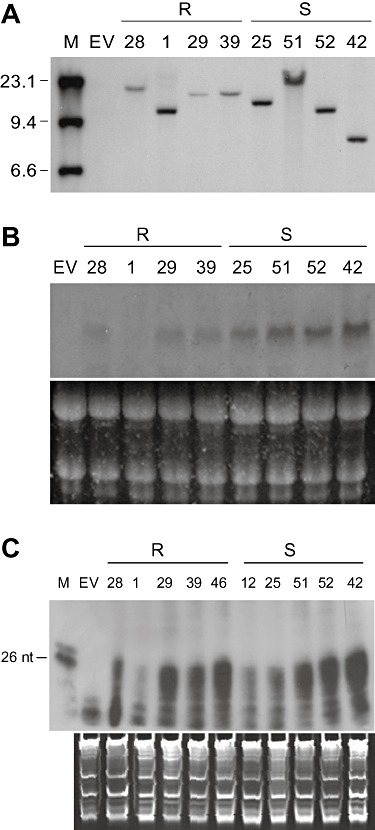
Analysis of nucleic acid preparations from non‐inoculated transgenic lime plants carrying a single integration of the intron‐hairpin 3′p23+3′UTR construct, the propagations of which were resistant (R) or susceptible (S) to Citrus tristeza virus (CTV) challenge. (A) Southern blot hybridization of DNA digested with ApaI, which cuts once at the left of the intron‐hairpin cassette, followed by electrophoresis in a 1% agarose gel and probing with a digoxigenin‐labelled DNA fragment derived from the Fad2 intron. M, digoxigenin‐labelled DNA molecular weight marker II (Roche). (B) Northern blot hybridization of the transgene‐derived transcript separated by electrophoresis in a formaldehyde‐containing 1% agarose gel and probed with a digoxigenin‐labelled DNA from the 3′p23+3′UTR construct. (C) RNase protection assay of the transgene‐derived siRNAs (as in Fig. 3B). M, radiolabelled DNA markers of 24 and 26 nucleotides. EV, nucleic acid preparation from control plants transformed with the empty vector. Equal RNA load was assessed by gel electrophoresis and ethidium bromide staining (B and C, bottom panels).
