Table 1.
Quantitative trait loci (QTLs) detected for each scoring date.
| Scoring date | QTL | MR* | KW† | CIM‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Closest marker | P value | Closest marker | K § | Df¶ | P value | Closest marker | LOD** | R 2 †† |
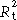 ‡‡
‡‡
|
||
| GP1 | PPV‐1H.a | ssrPaCITA05 | 3.5 × 10−3 | ||||||||
| GP2 | PPV‐1H.a | SSR03AG51 | 11.67 | 1 | 6.4 × 10−4 | ||||||
| GP2 | PPV‐1H.b | Cd195ssr | 4 × 10−4 | N86B11ssr3 | 11.75 | 1 | 6.1 × 10−4 | ||||
| GP2 | PPV‐1H.c | SSR01iso4G | 14.44 | 1 | 1.5 × 10−4 | SSR01iso4G | 2.84 | 7.15 | 26.90 | ||
| GP3 | PPV‐1H.a | pchcms4 | 32.83 | 1 | 1.0 × 10−8 | pchcms4 | 3.84 | 7.60 | 40.81 | ||
| GP3 | PPV‐1H.b | Cd195ssr | < 10−4 | N86B11ssr3 | 20.83 | 1 | 5.0 × 10−6 | N86B11ssr3 | 10.33 | 28.14 | |
| GP3 | PPV‐1H.c | UDAp‐414 | 23.61 | 1 | 1.2 × 10−6 | UDAp‐414 | 3.76 | 8.21 | |||
| GP3 | PPV‐3H.a | SSR02iso4G | 3.86 | 10.94 | |||||||
| GP4 | PPV‐1H.a | Cd195ssr | < 10−4 | pchcms4 | 47.02 | 1 | 7.0 × 10−2 | pchcms4 | 7.61 | 14.44 | 48.48 |
| GP4 | PPV‐1H.b | N86B11ssr3 | 37.21 | 1 | 1.1 × 10−9 | N86B11ssr3 | 15.17 | 39.44 | |||
| GP4 | PPV‐1H.c | SSR01iso4G | 23.29 | 1 | 1.4 × 10−6 | SSR01iso4G | 2.81 | 4.94 | |||
| GP5 | PPV‐1H.a | pchcms4 | < 10−4 | pchcms4 | 36.93 | 1 | 1.2 × 10−9 | pchcms4 | 5.25 | 12.59 | 41.44 |
| GP5 | PPV‐1H.b | N86B11ssr3 | 24.58 | 1 | 7.1 × 10−7 | ssrPaCITA05 | 10.79 | 33.26 | |||
| GP5 | PPV‐1H.c | UDAp‐414 | 13.47 | 1 | 2.4 × 10−4 | ||||||
| GP5 | PPV‐3H.a | SSR02iso4G | 3.4 × 10−3 | ||||||||
QTL detected in ‘Harlayne’ using QTLCartographer MultiRegress procedure.
QTL detected in ‘Harlayne’ using Kruskal–Wallis test.
QTL detected in ‘Harlayne’ using composite interval mapping.
Kruskal–Wallis test value.
Degree of freedom.
Logarithm of odds score (composite interval mapping).
Individual contribution of the QTL to phenotypic variance.
Total contribution of the detected QTL to phenotypic variance.
