Figure 3.
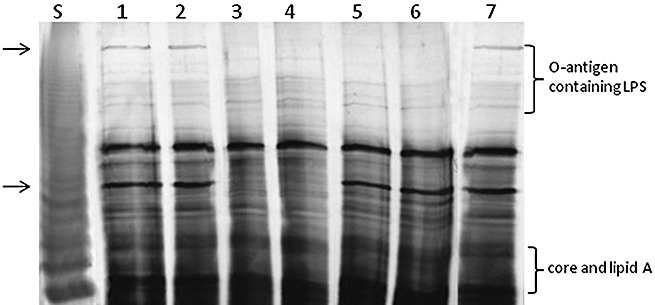
Sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of lipopolysaccharide (LPS) from proteinase K‐digested whole‐cell lysates of Xanthomonas citri ssp. citri strain 306 and its mutants carrying EZ‐Tn5 insertions in the wxacO and rfbC genes. Both wxacO and rfbC mutants were affected in the biosynthesis of O‐antigen. The wild‐type (Wt) LPS profile was restored in the complemented strains. S, LPS standard from Salmonella typhimurium (12.5 µg; Sigma). Lanes: 1, wild‐type strain 306; 2, C247C8 (wxacO mutant complemented with pJU3596); 3, 247C8 (wxacO) (wxacO mutant); 4: 247C8V (wxacO mutant complemented with empty vector pUFR053 without the wxacO gene); 5, 236E9 (rfbC) (rfbC mutant); 6, 236E9V (rfbC mutant complemented with empty vector pUFR053 without the rfbC gene); 7, C236E9 (rfbC mutant complemented with pJU3598). The gel was run in the SDS–glycine buffer system and silver stained using a silver stain kit (Bio‐Rad Laboratories, Inc.) following the manufacturer's instructions. The lost bands in the mutants are indicated by arrows.
