Figure 1.
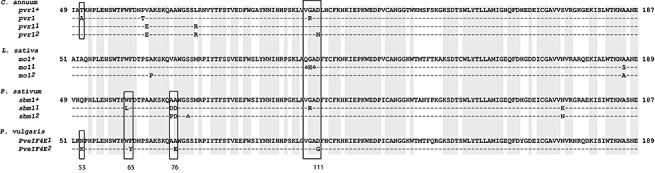
Comparison of amino acid polymorphisms in eukaryotic translation initiation factor 4E (eIF4E) found between susceptible and resistant/tolerant genotypes of Capsicum annuum (pvr1+, AY485127; pvr1, AY485129; pvr11, AY485130; pvr12, AY485131), Lactuca sativa (mo1+, AAP86602; mo11; mol2) and Pisum sativum (sbm1+, AAR04332; sbm1, AAT44121; sbm11, AAT44122) with eIF4E encoded by Phaseolus vulgaris alleles PveIF4E1 (EF571267) and PveIF4E2 (EF571275). PveIF4E2 found in the genotypes carrying the bc‐3 gene. The amino acids conserved in susceptible alleles of all four species are shaded in grey. Codons mutated in PveIF4E2 are indicated by numbers and similarly located mutations in eIF4E genes linked to potyvirus resistance in the other species are framed by rectangles. The symbol Δ indicates the amino acids deleted.
