Figure 1.
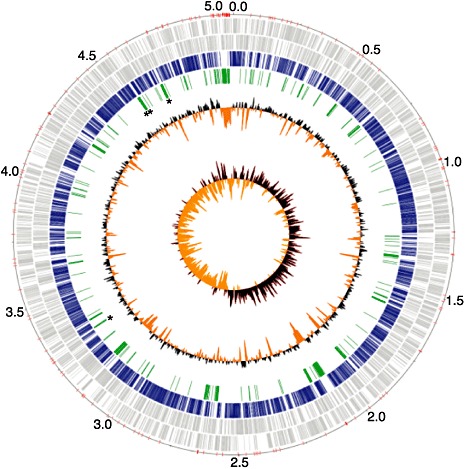
Circular representation of the improved, high‐quality draft genome sequence of Xanthomonas hortorum pv. carotae (Xhc) M081. Circle 1 (outer) designates the coordinates of the genome in half million base pair increments (red tick marks denote contig breaks); circles 2 and 3 show predicted coding sequences (CDSs) as grey lines on the positive and negative strands, respectively; circle 4 shows CDSs with homology to at least one other gene in other xanthomonads (e‐value <1 × 10−7); circle 5 shows regions of Xhc M081 larger than 1 kb with no detectable homology to genomes of other xanthomonads; asterisks highlight the locations for the Xhc‐specific primer sets XhcPP02 (between genome coordinates 3.0–3.5) and XhcPP03–XhcPP05 (in numerical order starting near genome coordinate 4.5; see also 2, 5 and Fig. 4); circle 6 shows deviations from the average GC percentage of 63.7% (black, greater; orange, smaller); circle 7 shows GC skew with bias for and against guanine as black and orange, respectively.
