Figure 1.
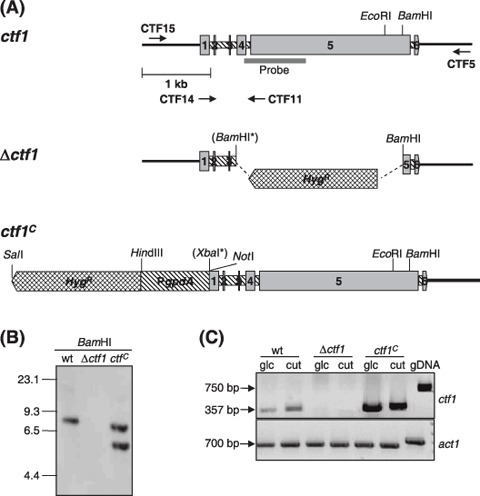
Targeted disruption and overexpression of the F. oxysporum ctf1 gene. (A) Physical maps of the ctf1 locus, the gene replacement vector (Δctf1 allele) and the ctf1C allele (ctf1 coding region fused to PgpdA from A. nidulans). The positions of the ctf1 probe, the HygR resistance cassette and the PgpdA promoter are indicated. The BamHI and XbaI sites introduced by site‐directed mutagenesis are marked with asterisks. (B) Southern hybridization analysis of wild‐type strain 4287 and transformants Δctf1 and ctf1C. Genomic DNA treated with BamHI was hybridized to the labelled ctf1 probe indicated in (A). The Δctf1 mutant shows homologous integration of the vector, whereas the constitutive ctf1C mutant carries an ectopic insertion of the ctf1C allele. Positions of DNA molecular size standards are shown. (C) Total RNAs from wild‐type, Δctf1 and ctf1C strains grown in SM with 1% glucose or 0.2% apple cutin as carbon sources were used for RT‐PCR analysis with ctf1‐specific primers. F. oxysporum genomic DNA was used as a control template. The actin gene (act1) was used as a control for equal amounts of RNA.
