SUMMARY
Efficient nutrient acquisition is critical to the fitness of plant pathogens. To address how the late blight agent Phytophthora infestans adapts to nutrients offered by its hosts, genes in glycolytic, gluconeogenic and amino acid pathways were mined from its genome and their expression in different plant tissues and artificial media was measured. Evidence for conventional glycolytic and gluconeogenic processes was obtained, although several steps involved pyrophosphate‐linked transformations which are uncommon in eukaryotes. In media manipulation studies, nearly all genes in the pathways were subject to strong transcriptional control. However in rye–sucrose media, tomato leaflets, potato tubers and, at both early and late stages of infection, most glycolytic genes were expressed similarly, which indicated that each plant tissue presented a nutrient‐rich environment. Biochemical analyses also demonstrated that sporulation occurred from host material in which sugars were abundant, with fructose and glucose increasing at the expense of sucrose late in the disease cycle. The expression of only a few genes changed late in infection, with the most notable example being lower invertase levels in the sucrose‐reduced leaves. Interestingly, most gluconeogenic genes were up‐regulated in tubers compared with other tissues. Rather than reflecting a starvation response, this probably reveals the role of such enzymes in converting carbon skeletons from the abundant free amino acids of tubers into citric acid cycle and glycolysis intermediates, as genes involved in amino acid catabolism were also more highly expressed in tubers. The corresponding enzymes also displayed higher activities in defined media when amino acids were abundant, as in tubers.
INTRODUCTION
The establishment of a profitable feeding relationship with a host is a prerequisite for the survival of most phytopathogenic fungi and oomycetes. To ensure an efficient interaction, pathogen metabolism needs to adapt to the nutrients offered by a host, including changes that transpire during the course of infection. Perception of nutrient concentrations by the pathogen may also be needed to elicit certain developmental stages, such as haustoria for feeding or spores for dispersal. Although much has been learned about the in vitro nutritional requirements of some fungi and how their transcriptomes adapt to nutrients in artificial media, the situation within infected plants remains poorly studied (Divon and Fluhr, 2007; Gancedo, 1998; Solomon et al., 2003; Wong et al., 2008). Even less is known about the nutritional responses of oomycetes. Learning how nutrients are acquired and metabolized by such species may be useful for developing novel control strategies and for understanding interactions between disease, agronomic procedures, such as fertilization, and other environmental factors.
Most fungi and oomycetes rely on stored nutrients during the initial stages of infection, but start to utilize host compounds after penetration. The major plant nutrients used include sugars and amino acids, which are typically mobilized using membrane‐bound transporters on hyphae or haustoria (Mendgen and Hahn, 2002). There is no simple answer as to whether colonization sites in plants are perceived by pathogens as having abundant or limiting nutrients, but some insight has resulted from the study of the expression of metabolic genes of the pathogen in planta. Such work has suggested that many plants are nutrient‐rich, although this may vary between pathosystems and the stage of infection (Bolton and Thomma, 2008; Keon et al., 2007). In addition, some pathogens are believed to directly or indirectly stimulate hosts to transport photosynthate towards infection sites (Abood and Losel, 2003; Doehlemann et al., 2008).
Although assays of plant contents may indicate the types and quantities of potential nutrients, it is unclear which are used preferentially by pathogens. Such analyses are also complicated by the fact that nutrients may change throughout the day, growing season or infection time course. Variation also occurs between species and organs on the same plant, such as foliage, tubers, roots and fruit. Although it would be in the best interest of a pathogen to tailor its metabolism to each host situation, the extent to which this occurs is unknown. The degree of flexibility in metabolic adaptation may help to explain why some species are cosmopolitan, host‐specific or organ‐specific pathogens.
The oomycete Phytophthora infestans colonizes the stems and leaves of potato and tomato, plus potato tubers and tomato fruit, causing the disease called late blight. These organs offer distinct environments to the pathogen as a result of their varied physiologies and cellular structures. In both types of tissue, P. infestans establishes intercellular and intracellular hyphae and haustoria through which most nutrient uptake is thought to occur (Hohl and Stoessel, 1976; Hohl and Suter, 1976). However, it is unknown whether the metabolic strategies employed by P. infestans vary during growth in its different hosts. To address this, the expression of pathogen genes associated with glycolysis, gluconeogenesis and amino acid metabolic pathways was measured during the infection of tomato leaves and potato tubers. Significant differences were observed between tuber and leaf infections, especially for genes involved in gluconeogenic and anapleurotic pathways. Infection was also associated with an increase in free hexoses at the expense of sucrose and/or starch.
RESULTS
Annotation of carbohydrate metabolism genes
As a first step towards the analysis of the expression of metabolic genes in P. infestans during infection, the genome database developed by the Broad Institute (http://www.broad.mit.edu) was surveyed for participants in glycolysis and gluconeogenesis. This entailed keyword searches and iterative rounds of BlastP using non‐oomycete and Phytophthora query sequences. Gene models were evaluated using expressed sequence tags, proteomic data and alignments to P. ramorum and P. sojae. Models were corrected as needed and uploaded to the database.
The analysed genes are listed in Table 1. In addition to genes directly related to glycolysis and gluconeogenesis, several that were indirectly related were also included. The latter included: invertase, as sucrose is a likely entry point for sugars into glycolysis during infection; malate dehydrogenase, which transports mitochondrial oxaloacetate to the cytoplasm for use in gluconeogenesis; 6‐phosphofructo‐2‐kinase (fructose‐2,6‐bisphosphatase), which represses the allosteric enzyme 6‐phosphofructose‐1‐kinase, which is a key regulator of glycolysis; and pyruvate phosphate dikinase, which can form the gluconeogenic intermediate phosphoenolpyruvate. The latter enzyme, which has been identified previously in Phytophthora, is of evolutionary interest as it is largely limited to plants, bacteria and selected protists (Marshall et al., 2001; Slamovits and Keeling, 2006). In addition to generating phosphoenolpyruvate, pyruvate phosphate dikinase may also convert phosphoenolpyruvate, AMP and pyrophosphate (PPi) into pyruvate and ATP as a bypass to pyruvate kinase in glycolysis.
Table 1.
Number of genes in glycolytic and other pathways.
| Pathway and enzymatic activity* | Symbol | Number of predicted genes† | |||||
|---|---|---|---|---|---|---|---|
| P. infestans | P. ramorum | Yeast | Arabidopsis | Diatom | Human | ||
| Glycolysis | |||||||
| Invertase (EC 3.2.1.26) | INV | 3 | 3 | 6 | 11 | 0 | 1 |
| Hexokinase (EC 2.7.1.2) | HXK | 5‡ | 4 | 4 | 6 | 1 | 4 |
| Glucose‐6‐phosphate isomerase (EC 5.3.1.9) | GPI | 1 | 1 | 1 | 2 | 2 | 2 |
| 6‐Phosphofructokinase (EC 2.7.1.11) | FPK | 1 | 1 | 2 | 7 | 2 | 3 |
| Fructose‐bisphosphate aldolase (EC 4.1.2.13) | ALD | 3 | 3 | 1 | 2 | 4 | 3 |
| Triosephosphate isomerase (EC 5.3.1.1) | TPI | 2 | 2 | 1 | 2 | 4 | 3 |
| TPI + GPD gene fusion | TPI‐GPDH | 1 | 1 | 0 | 0 | 2 | 0 |
| Glyceraldehyde‐3‐phosphate dehydrogenase (EC 1.2.1.12) | GPDH | 7‡ | 6 | 3 | 8 | 3 | 2 |
| Phosphoglycerate kinase (EC 2.7.2.3) | PGK | 2 | 2 | 1 | 3 | 5 | 1 |
| Phosphoglycerate mutase (EC 5.4.2.1)§ | PGM | 2 | 2 | 3 | 3 | 10 | 2 |
| Enolase (phosphopyruvate hydratase; EC 4.2.1.11) | ENO | 4 | 3 | 2 | 3 | 2 | 4 |
| Pyruvate kinase (EC 2.7.1.40) | PYK | 6‡ | 7 | 2 | 14 | 5 | 2 |
| Gluconeogenesis‐related | |||||||
| Pyruvate carboxylase (EC 6.4.1.1) | PYC | 1 | 1 | 2 | 3 | 2 | 1 |
| Malate dehydrogenase (EC 1.1.1.37) | MDH | 2 | 2 | 3 | 10 | 3 | 1 |
| Phosphoenolpyruvate carboxykinase (EC 4.1.1.49) | PEPCK | 4 | 4 | 1 | 2 | 1 | 0 |
| Fructose‐1,6‐bisphosphatase (EC 3.1.3.11) | FBP | 1 | 2 | 1 | 3 | 3 | 2 |
| Other | |||||||
| Pyruvate, phosphate dikinase (EC 2.7.9.1) | PPDK | 2 | 2 | 0 | 1 | 1 | 0 |
| 6‐Phosphofructose‐2‐kinase (EC 2.7.1.105)¶ | PFK2 | 3 | 3 | 2 | 7 | 1 | 4 |
Some enzymes may also function in other pathways.
Number of predicted genes from Phytophthora, Saccharomyces (yeast), Arabidopsis thaliana, Thalassiosira pseudonana (diatom) and Homo sapiens (human). Phytophthora genes were manually annotated, whereas those in the other genomes were taken from their public assemblies.
Includes one apparent pseudogene.
Does not include the seven apparent non‐glycolytic mutases.
Bifunctional enzyme that also contains fructose‐2,6‐bisphosphate 2‐phosphatase activity (EC 2.7.1.11).
Although addressing the role of pyruvate phosphate dikinase in P. infestans is complicated by its coexistence with the ATP‐dependent pyruvate kinase, other steps in glycolysis also appear to be PPi‐based. For example, only a PPi‐dependent 6‐phosphofructose‐1‐kinase was found, as opposed to the ATP‐dependent form of animals and many bacteria. However, a PPi‐dependent phosphoenolpyruvate carboxytransphosphorylase (E.C. 4.1.1.38) which, in Entamoeba, substitutes for the traditional phosphoenolpyruvate carboxykinase (E.C. 4.1.1.49) in the generation of oxaloacetate was not detected in P. infestans.
As plant fructose may be important to P. infestans metabolism during infection, in addition to sucrose and glucose, the genome was checked for specialized fructokinases (E.C. 2.7.1.4) typical of those found in plants. None were detected. Hexokinases may therefore phosphorylate fructose for entry into glycolysis, as in animals.
Organization of gene families
Of the 18 activities shown in Table 1, five are encoded in P. infestans by a single gene and the rest by families. For example, one gene is predicted for glucose‐6‐phosphate isomerase and five for pyruvate kinase. Although much variation exists between oomycetes and non‐oomycetes in the number of genes for each enzyme (Table 1), P. ramorum and P. infestans typically have equal quantities. The exceptions are fructose‐1,6‐bisphosphatase, for which P. infestans and P. ramorum appear to contain one and two genes, respectively, and pyruvate kinase, for which five and seven are predicted. Phytophthora infestans also appears to contain hexokinase, glyceraldehyde‐3‐phosphate dehydrogenase and pyruvate kinase pseudogenes.
Members of families are often physically linked. For example, clusters are observed for all three invertases, all five hexokinases, two of three fructose‐1,6‐bisphosphate aldolases, two of six glyceraldehyde phosphate dehydrogenases and both pyruvate phosphate dikinases. The sizes of the regions spanned by the clusters vary dramatically. For example, although the two pyruvate phosphate dikinases are 2 kb from each other, the five hexokinases are spread over 27 kb and the three invertases over 73 kb. Most clusters observed in P. infestans were also detected in P. ramorum.
The sizes of most gene families are different in the diatom Thalassiosira pseudonana and Phytophthora species, even though are all classified as Stramenopiles. Some of this variation is probably attributable to their autotrophic vs. heterotrophic lifestyles. For example, the two Phytophthora species express three invertases and four active hexokinases, which are probably important for the utilization of plant sugars. In contrast, the diatom lacks invertase and contains only one hexokinase, which presumably reflects its use of the primary products of photosynthesis for energy.
A previous study has reported that the genes encoding triosephosphate isomerase and glyceraldehyde‐3‐phosphate dehydrogenase are fused into a single transcriptional unit in P. infestans (Unkles et al., 1997). Based on hybridization data, this study concluded that independent triosephosphate isomerase genes did not exist. However, two and six unfused triosephosphate isomerase and glyceraldehyde‐3‐phosphate dehydrogenase genes, respectively, were detected in both P. infestans and P. ramorum. A combination of fused and non‐fused loci also occurs in the diatom.
Gene expression in planta and in vitro
Levels of mRNA for each gene were measured during growth in rye–sucrose media, tomato foliage and potato tubers. To ensure that equivalent developmental stages were analysed, tissues were harvested on the first day on which sporulation was observed, thereby representing the completion of one disease or life cycle. This corresponded to 5‐day‐old rye–sucrose agar cultures, and 6‐day post‐infection tuber slices and tomato leaves. Samples from tomato 3 days after inoculation were also examined. For each treatment, two independent biological replicates were prepared in different weeks. For each replicate, RNA was pooled from a minimum of four rye–sucrose cultures, five plants or two slices from each of four different tubers.
When quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assays were performed against the 50 genes in Table 1, reliable signals were detected against 46 genes. These are reported in Fig. 1B, in which the enzymes are listed in the same order as in Table 1 and named using their gene model identifier from the Broad Institute (PITG_XXXXX). Values were normalized to ribosomal protein S3a, which is expressed at similar levels throughout the life cycle (Judelson et al., 2008; Yan and Liou, 2006). One gene not shown is the glyceraldehyde‐3‐phosphate dehydrogenase locus PITG_01940, as gene‐specific primers could not be developed because of its similarity to PITG_01938; the results presented for the latter therefore include both genes. Other genes not shown in Fig. 1 lacked detectable transcripts. These included the apparent pseudogenes for glyceraldehyde‐3‐phosphate dehydrogenase (PITG_19495) and pyruvate kinase (located near base 994 250, scaffold 40). In contrast, transcripts were detected for the hexokinase pseudogene PITG_06022.
Figure 1.
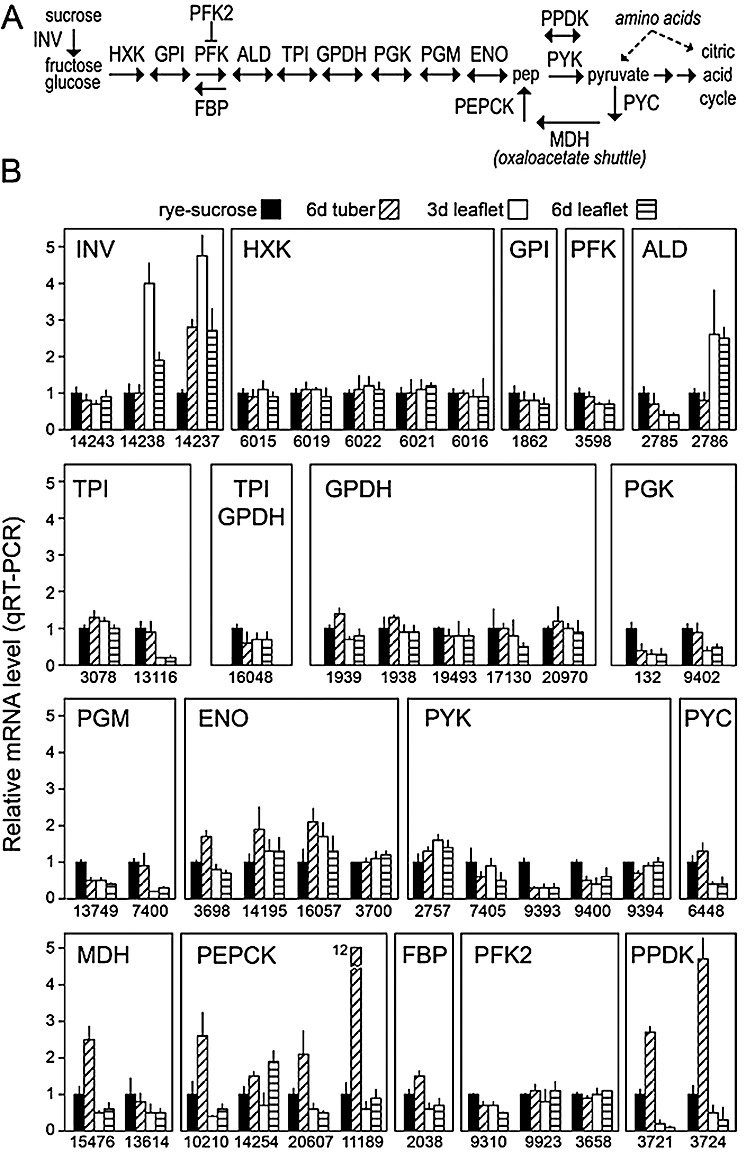
(A) Diagram of metabolic pathways illustrating the role of the enzymes involved in glycolysis and gluconeogenesis. Enzymes are shown in capital letters using the abbreviations given in Table 1, and selected compounds and other pathways are presented in lower case letters. (B) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of the genes. RNA was extracted from rye–sucrose cultures, potato tubers 6 days after infection and tomato leaflets 3 and 6 days after infection, as indicated. Assays were performed in quadruplicate, including two biological replicates. mRNA levels were normalized using the Phytophthora infestans gene for ribosomal protein S3a, and are reported relative to the concentration in rye–sucrose cultures (y‐axis). Shown at the base of each figure are gene identifiers from the Broad Institute database (PITG_XXXXX).
The most striking pattern in the data was the up‐regulation of most gluconeogenesis‐related genes in potato tubers, compared with rye–sucrose media and tomato leaflets. These included all pyruvate carboxylases, phosphoenolpyruvate carboxykinases, fructose‐1,6‐bisphosphatases and pyruvate phosphate dikinases, although the magnitudes of induction varied. For example, pyruvate carboxylase PITG_06448 was induced 30% in tubers compared with the other tissues, the phosphoenolpyruvate carboxykinase PITG_11189 was induced 12‐fold, and the other phosphoenolpyruvate carboxykinases were induced about two‐fold. In contrast, most gluconeogenesis‐associated mRNAs were found at similar levels in rye–sucrose media and tomato, or showed lower levels in tomato. For example, pyruvate carboxylase was down‐regulated three‐fold in tomato compared with rye–sucrose media.
The majority of glycolytic genes varied little between tissues, compared with the relatively consistent changes in gluconeogenesis‐associated loci. For example, all members of the hexokinase and glyceraldehyde‐3‐phosphate dehydrogenase families exhibited similar mRNA levels in rye–sucrose media, tomato and potato. In addition, glucose‐6‐phosphate isomerase and 6‐phosphofructokinase genes were expressed at similar levels in rye–sucrose media and tubers, and at similar or slightly lower levels in tomato.
A few glycolysis‐associated genes exhibited significant changes, although divergence existed among different members of each enzyme family. For example, only two of the three invertases were up‐regulated in tomato leaflets and/or potato tubers compared with rye–sucrose cultures. In addition, although the aldolase PITG_02785 was down‐regulated in tomato compared with rye‐sucrose media and tubers, the PITG_02786 form of this enzyme was up‐regulated in tomato.
Tomato samples at both 3 and 6 days post‐infection were analysed to check whether a change in nutrient concentration during infection altered gene expression. For most genes, mRNA levels were similar at the two time points. The major exceptions were the invertases PITG_14237 and PITG_14238, for which the mRNA levels dropped by one‐half late in infection. This may be explained by the drop in sucrose observed at the late time point, as described later. In addition, the phosphoenolpyruvate carboxykinase locus PITG_14254 showed higher mRNA levels late in infection.
To confirm the qRT‐PCR data, five genes were analysed using RNA blots. This focused on genes that were up‐regulated in tubers compared with rye–sucrose media and tomato. The results are presented in Fig. 2, together with a quantitative comparison of the RNA blot and qRT‐PCR data. Good agreement was observed between the two methods.
Figure 2.
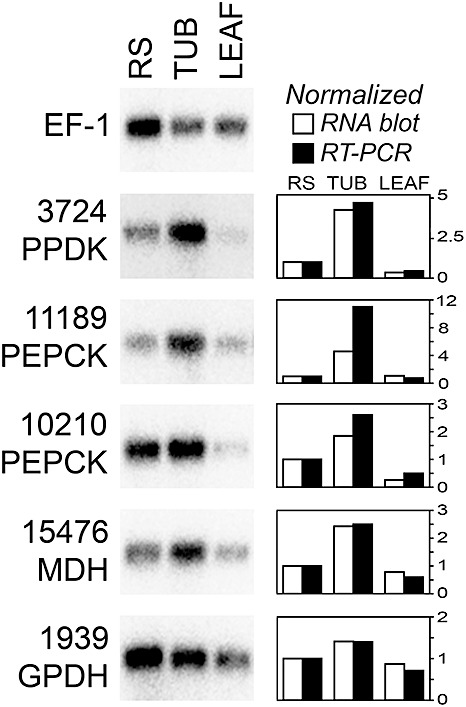
RNA blot analysis of selected carbohydrate metabolism genes. RNA was from rye–sucrose cultures (RS), tubers 6 days post‐infection (TUB) or tomato leaflets 6 days post‐infection (LEAF). To help adjust for different levels of Phytophthora infestans RNA, about 10 times more RNA was loaded for the two plant samples, and then a gene for P. infestans elongation factor 1‐α was hybridized as a control. Shown at the right side of the figure are quantitative data for each gene. This was determined by phosphorimager analysis of the RNA blot, or quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) as shown in Fig. 1.
Sensitivity of mRNA levels and enzyme activities to sucrose
The lack of major changes in most glycolysis‐regulated mRNAs was unanticipated, as it was assumed that the different growth conditions would present P. infestans with divergent nutrient levels. In addition, in other organisms, such genes are frequently sensitive to fluctuating nutrients (Moore et al., 1991; Nakajima et al., 2000). To assess the ability of P. infestans to regulate the genes, their expression in low‐ and high‐sucrose media was compared. This involved growing hyphae in rye broth for 4 days without added sucrose, and then providing 2% sucrose for 2 h. As rye broth (made from 1‐day germinated grain) contains 0.08% sucrose and smaller amounts of fructose and glucose, the sucrose pulse would therefore increase soluble sugars by 20‐fold.
Based on qRT‐PCR, most of the 46 genes exhibited dramatic changes in expression in response to the sucrose pulse (Fig. 3). For example, the addition of sucrose increased the mRNA levels of the three invertases by more than four‐fold. This suggests that P. infestans has a strong ability to respond to sucrose, which is the most abundant soluble sugar in typical plant tissues and is a probable source of most carbon entering glycolysis.
Figure 3.
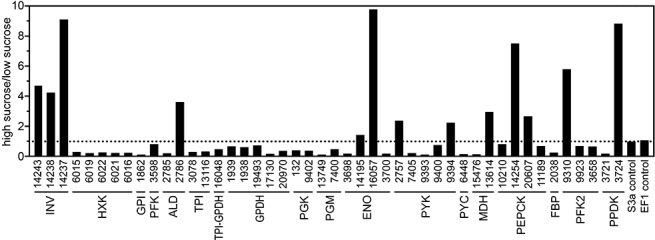
Expression of carbohydrate metabolism genes in response to sucrose. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed using mRNA from rye broth cultures of Phytophthora infestans or rye broth cultures subjected to a 2‐h pulse of 2% sucrose. Assays were performed in quadruplicate, including two biological replicates. mRNA levels were normalized using the P. infestans gene for ribosomal protein S3a, and are reported as a ratio of the levels in high‐sucrose vs. low‐sucrose conditions; the broken line represents no change between the treatments.
In contrast with the invertases, nearly all glycolytic genes were down‐regulated by the sucrose pulse. For example, mRNAs for the six hexokinases fell by an average of five‐fold, and glucose phosphate isomerase mRNA dropped 10‐fold. However, a few glycolytic genes responded differently. For example, although the PITG_2785 aldolase was down‐regulated by the added sucrose, the PITG_2786 aldolase was up‐regulated. Heterogeneous responses were also observed within the enolase and pyruvate kinase families.
Genes for two key enzymes involved in gluconeogenesis, pyruvate carboxykinase and fructose‐1,6‐bisphosphatase, were also repressed by the sucrose pulse. As these enzyme activities are encoded by single genes within P. infestans, their down‐regulation may indicate an effort to reduce flux through the pathway. An alternative explanation for the down‐regulation of pyruvate carboxykinase (which converts pyruvate to oxaloacetate) may be a reduced need to replenish citric acid cycle intermediates. The latter may also be related to the induction by sucrose of PITG_13614 mRNA, which encodes the mitochondrial form of malate dehydrogenase; this aids in the removal of oxaloacetate from the mitochondria by converting it to malate, which can be transported across the mitochondrial membrane.
Different members of gene families encoding many enzymes involved in gluconeogenesis showed divergent responses. For example, the sucrose pulse caused mRNAs for the mitochondrial malate dehydrogenase to rise and, mRNAs for its cytoplasmic form, PITG_15476, to fall. However, although sucrose caused mRNAs for only some of the phosphoenolpyruvate carboxykinases to increase, the changes did not correlate with their subcellular localization; PITG_10210 and PITG_20607 are predicted to be mitochondrial, whereas PITG_14254 and PITG_11189 are cytoplasmic.
To confirm the qRT‐PCR data, five genes were analysed using RNA blots. As shown in Fig. 4, strong agreement was observed between the two methods. For example, both indicated that sucrose caused mRNA from PITG_03724 to rise (pyruvate phosphate dikinase) and mRNA from PITG_15476 to fall (malate dehydrogenase).
Figure 4.
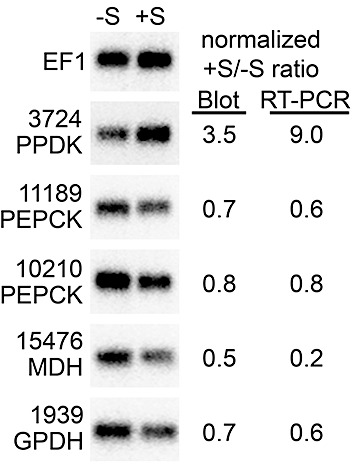
RNA blot analysis of selected carbohydrate metabolism genes from sucrose‐amended and unamended broth cultures. RNA was taken from the same samples as used in Fig. 3 (–S, rye broth; +S, 2‐h sucrose pulse in rye broth). Shown on the right side of the figure are quantitative data for each gene from phosphorimager analysis of the blot, or quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) data.
To test whether the changes in mRNA abundance shown in Fig. 3 were reflected in alterations of the corresponding enzyme activities, four enzymes were assayed in extracts of plain rye medium and sucrose‐treated samples (Fig. 5). The results were consistent with the findings from qRT‐PCR. For example, invertase activity increased 2.5‐fold during the sucrose pulse. This is lower than the four‐ to nine‐fold increase in mRNA observed for the corresponding genes, but a greater rise in enzyme would probably have been observed if the sucrose pulse had been longer than 2 h. Similarly, the sucrose pulse caused the hexokinase activity to diminish by three‐fold, compared with an average decrease of five‐fold in mRNA from the five hexokinase genes. Malate dehydrogenase and phosphoenolpyruvate carboxykinase activities showed minor or no changes, which is consistent with the observation that some of their corresponding genes rose and others fell in response to the sucrose pulse.
Figure 5.
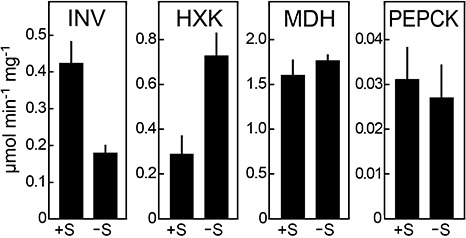
Enzyme activities from sucrose‐amended and unamended broth cultures. Total invertase (INV), hexokinase (HXK), malate dehydrogenase (MDH) and phosphoenolpyruvate carboxykinase (PEPCK) activities were measured in extracts from hyphae grown in plain rye both (–S) or after the 2‐h sucrose pulse (+S). Activities are the means of two biological replicates, and are expressed as specific activity (micromoles of substrate converted per minute per milligram of protein).
Carbohydrate levels during infection
Fructose, glucose, sucrose and starch within plant tissues were measured during colonization by P. infestans to place the expression of the metabolic genes in context with the available carbohydrates (Fig. 6). At the onset of infection, tubers showed higher levels of fructose [19 vs. 6 µmol/g fresh weight (FW)] and glucose (16 vs. 4 µmol/g FW) than did tomato leaves. Sucrose concentrations were more equivalent (16 vs. 12 µmol/g FW). These values are within the ranges described previously for such tissues (Berger et al., 2004; Bialczyk et al., 2005; Coffin et al., 1987; Daniele et al., 2003). By comparison, rye–sucrose medium contains 53 µmol/g wet weight of sucrose and less than 0.3 µmol/g of fructose and glucose.
Figure 6.
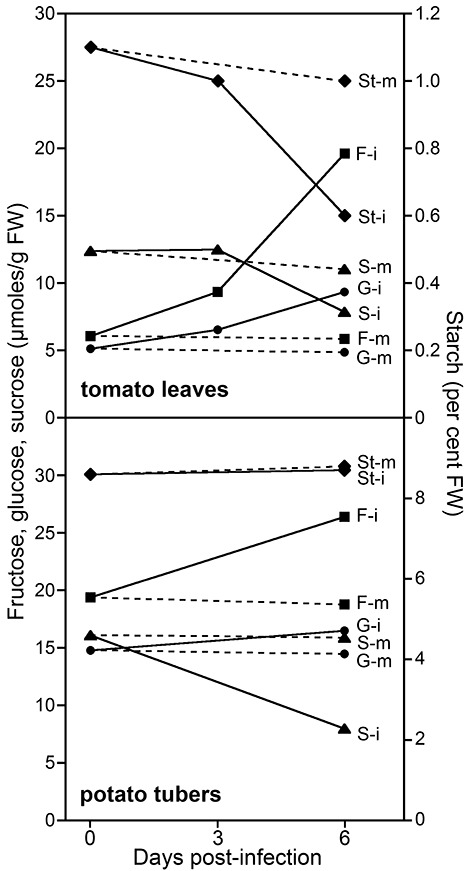
Effect of infection on carbohydrate levels in leaves and tubers. Concentrations of fructose (F, squares), glucose (G, circles), sucrose (S, triangles) and starch (St, diamonds) were measured in infected (i) and mock‐infected (m) tomato leaves and tubers at the indicated times after inoculation with zoospore suspensions. Multiple infected samples were pooled and then assayed in duplicate. Error bars are omitted for clarity.
Substantial changes in carbohydrate were observed over the course of the 6‐day infection. In tomato leaves, fructose levels increased three‐fold and glucose doubled, whereas starch and sucrose fell by one‐half. Only minor changes were observed in mock‐infected samples. The increase in free hexoses at the expense of sucrose in the infected tomato leaves is consistent with changes described in potato leaves (Daniele et al., 2003; Engstrom and Stromberg, 1996), which is presumably a result of increases in host invertases (Berger et al., 2004; Biemelt and Sonnewald, 2006; Roitsch et al., 2003).
Compared with the situation in tomato leaves, a smaller increase in free hexoses was observed in infected tubers. Fructose concentrations increased by about one‐third, glucose rose slightly and sucrose dropped by one‐half. All three soluble sugars decreased slightly in the mock‐infected tubers.
Amino acid metabolism in infected plants
The abundance of sugars in potato tubers suggested that the up‐regulation of mRNAs for most gluconeogenic enzymes in this tissue was not a result of sugar deprivation. Therefore, the possibility was considered that carbon entering from another source was being used to generate six‐carbon sugars (for example, for cell wall biogenesis) or to replenish citric acid cycle intermediates that would feed into other pathways (for example, for lipid synthesis). Amino acids are a strong candidate for this alternative carbon source. Free amino acids are very abundant in tubers, typically accounting for 1%–1.5% of fresh weight, where they represent about one‐half of total amino acids. The concentration of free amino acids in tubers is 10‐fold higher than in tomato or potato leaves, where most amino acids are within proteins (Bialczyk et al., 2005; Brierley et al., 1996; Carpena‐Ruiz et al., 1989).
To explore the involvement of amino acids, several P. infestans genes related to their metabolism were assayed in the rye–sucrose, tomato and tuber samples by qRT‐PCR (Fig. 7). The liberation of carbon skeletons from most amino acids involves cytoplasmic transaminases that shift the amino groups to α‐ketoglutarate, forming glutamate. Glutamate then undergoes oxidative deamination in the mitochondria by glutamate dehydrogenase to reform α‐ketoglutarate. Therefore, both glutamate dehydrogenases and several transaminases were examined.
Figure 7.
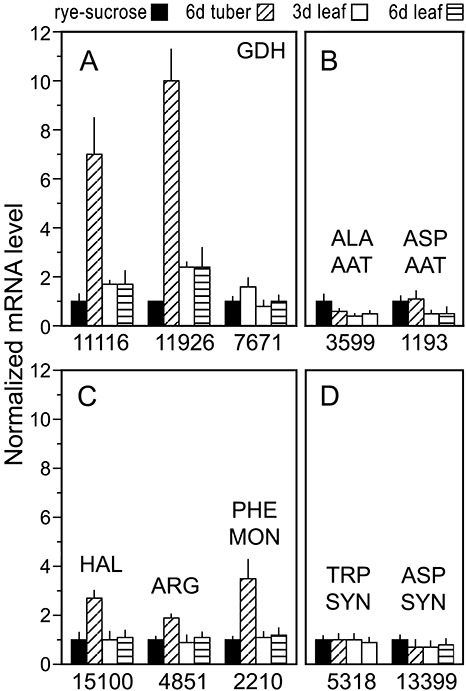
Expression of amino acid metabolism genes in laboratory media, infected tubers and leaves. RNA was analysed as in Fig. 1. (A) Glutamate dehydrogenases (GDH); (B) alanine and aspartate aminotransferases (ALA AAT, ASP AAT); (C) histidine ammonia lyase (HAL), arginase (ARG) and phenylalanine monooxygenase (PHE MON); (D) tryptophan and aspartate synthases (TRP SYN, ASP SYN).
Three genes encoding glutamate dehydrogenase (E.C. 1.4.1.2) were found in the genome, and all exhibited higher mRNA levels in tubers compared with rye–sucrose cultures or tomato leaflets (Fig. 7A). Two were also slightly higher in tomato than in rye–sucrose media. However, mRNA levels for two transaminases showed little difference between the samples, or slightly higher levels in rye–sucrose media and tubers than tomato (Fig. 7B). These were alanine aminotransferase (E.C. 2.6.1.21) and aspartate aminotransferase (E.C. 2.6.1.1).
As glutamate dehydrogenases and transaminases catalyse reversible reactions, their expression is an imperfect marker for amino acid catabolism. Consequently, three enzymes that play specific roles in amino acid breakdown were analysed. These were histidine ammonia lyase (E.C. 4.3.1.3), which converts histidine to urocanate, arginase (E.C. 3.5.3.1), which degrades arginine to ornithine and urea, and phenylalanine monooxygenase (E.C. 1.14.16.1), which is the first step in phenylalanine degradation. All three showed significantly higher mRNA levels in tubers than in rye–sucrose media or tomato (Fig. 7C), supporting the hypothesis that P. infestans utilizes the free amino acids of tubers as a major carbon source.
As a control, mRNA levels for two enzymes specifically involved in amino acid biosynthesis were examined. These were tryptophan synthase (E.C. 4.2.1.20) and asparagine synthase (E.C. 6.3.5.4). Both had similar mRNA levels in rye–sucrose, tuber and tomato leaflet samples (Fig. 7D).
Sensitivity of pathways to amino acids in vitro
To test further the premise that P. infestans adapts its metabolism to amino acids in its environment, four enzymes that varied between growth in leaflets and tubers were examined in cultures containing no or high levels of amino acids. This utilized the defined medium described by Xu (1982), which contains ammonium sulphate as the sole nitrogen source, and the same supplemented with 1% casamino acids. The mRNA levels of the four genes were determined by qRT‐PCR, and enzyme activities were measured using spectrophotometric assays (Fig. 8).
Figure 8.
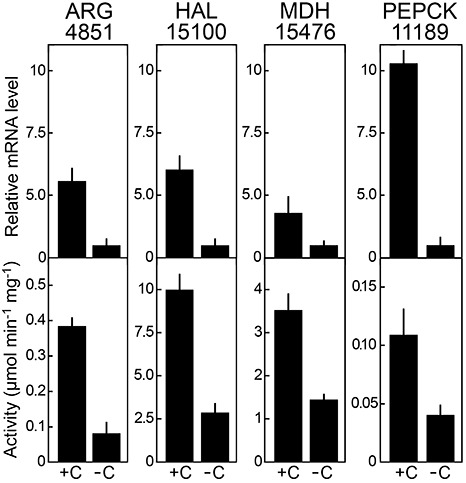
Effect of amino acids on enzyme expression. Hyphae were grown on defined media containing ammonium sulphate as the sole nitrogen source (–C) or supplemented with 1% casamino acids (+C). Top panels show mRNA levels as determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) for the arginase and histidine ammonia lyase genes, and for malate dehydrogenase PITG_15476 and phosphoenolpyruvate carboxykinase PITG_11189 genes; values were normalized using ribosomal protein S3a and then adjusted so that the –C sample equalled unity. Values are derived from analyses of two biological replicates, with two technical replicates per sample. Bottom panels show the specific activity of each enzyme, based on the two biological replicates.
The results were consistent with the hypothesis that gluconeogenic and amino acid catabolic pathways are up‐regulated in tubers as a result of amino acid abundance. The catabolic enzymes arginase and histidine ammonia lyase, for example, showed higher levels of both mRNA and enzymatically active protein during growth in the media supplemented with amino acids. The gluconeogenic enzyme phosphoenolpyruvate carboxykinase and the oxaloacetate‐shuttling enzyme malate dehydrogenase were also up‐regulated in the high‐amino‐acid media, paralleling the situation in tubers, at both the mRNA and protein level. It should be noted that phosphoenolpyruvate carboxykinase and malate dehydrogenase are encoded by gene families and, although the qRT‐PCR shown in Fig. 8 measured only single genes for each (PITG_11189 and PITG_15476), the others presumably also contributed to the enzyme activity. Arginase and histidine ammmonia lyase are single‐copy.
DISCUSSION
Organisms frequently adapt their metabolism to their environment. This is often observed when a preferred substrate is available as a carbon source, in which case the appropriate catabolic enzymes are expressed at higher levels. Conversely, alternative pathways are up‐regulated when that compound is limiting. In both cases, the fine‐tuning of central metabolism helps to yield the required balance of carbon‐based biosynthetic intermediates, and reduce futile cycling between pathways such as glycolysis and gluconeogenesis. Studies in industrial and model fungi have led to a fair understanding of carbon and nitrogen catabolite repression, the role of G‐protein‐coupled receptors in nutrient perception and how metabolism is integrated with development (Gancedo, 1998; Lorenz et al., 2000; Wong et al., 2008). Some work has also been performed in animal pathogens (Barelle et al., 2006; Hu et al., 2008). In contrast, much remains to be learned about how pathogens respond to nutrients in plants, which nutrients pathogens prefer and how nutrients impact on disease. Some metabolic genes are known to be pathogenicity factors (Idnurm and Howlett, 2001; Wei et al., 2004).
Our studies of P. infestans on diverse hosts and media confirm that oomycetes regulate their metabolism in response to nutrients in both artificial media and in planta. Although an altered mRNA level for an enzyme does not prove that flux through that step changes, and post‐translational control must also be considered, our data provide insight into the metabolic strategies of P. infestans. For example, the higher expression of enzymes in amino acid catabolism and gluconeogenesis within tubers suggests that amino acids may be a favoured carbon source. The average potential energy yield from an amino acid molecule is lower than that from glucose, 28 vs. 38 ATP (Ferrer‐Lorente et al., 2007). However, as a result of the high concentration of free amino acids in tubers, about 150 µmol/g FW, they yield more oxidizable carbon than do soluble sugars in this tissue (Brierley et al., 1996). More carbon is stored in tubers as starch, but, as this is sequestered in the amyloplast, it is presumably inaccessible to P. infestans.
Few changes in mRNA levels were observed between the 3‐ and 6‐day‐infected tomato leaf samples. This indicates that nutrient shifts over a disease cycle are insufficient to alter the transcription of most genes, and that, overall, nutrient limitation is not a major signal for sporulation. The most obvious change occurring at the later stage of infection involved two of the invertases, which showed lower mRNA levels at day 6, when sucrose levels had declined by one‐half. Although reduced sucrose may have been sufficient to down‐regulate the invertases, this is unlikely to have been perceived as a diminution in available carbon as fructose and glucose rose concomitantly. In addition, the hexoses are probably the primary sugar source for Phytophthora as its cell wall‐bound invertase activity substantially exceeds the maximum uptake rate of sucrose through transmembrane carriers (Sheard and Farrar, 1987).
The most likely cause of the fall in sucrose in both tubers and leaves is the induction of plant invertases, reflecting a shift from normal metabolism to defence. Although not described previously for P. infestans tuber disease, the induction of invertases is a general response in many pathosystems (Berger et al., 2004; Biemelt and Sonnewald, 2006; Roitsch et al., 2003). Whether the resulting higher levels of glucose and fructose are exploited by P. infestans to serve its own metabolic needs is a matter of conjecture.
Not all of the changes in mRNA levels are easily interpreted, particularly when gene families are involved. For example, the two fructose‐1,6‐bisphosphate aldolases responded divergently to the sucrose pulse, and between growth in rye–sucrose media and in planta. As members of most families are clustered, it might at first appear surprising that expression varies. In the case of the aldolases (PITG_02785 and PITG_02786), their coding regions are also 97% identical at the DNA level. However, there is no significant similarity between their promoters, which are separated by 16.4 kb. Several DNA and RNA transposons exist in the intervening region, which may have helped to diversify their expression.
The divergent expression of isoforms of carbohydrate‐metabolizing enzymes has also been well described in other taxa, where its physiological significance is also poorly understood (Ferris and Whitt, 1979; Lebherz and Rutter, 1969). Isoforms may, in theory, vary in substrate specificity, favour forward or reverse reactions, be linked to development or even participate in roles unrelated to metabolism. For example, in Saccharomyces cerevisiae, enolase and aldolase regulate vacuolar assembly and vacuolar ATPases in addition to their roles in glycolysis (Gancedo and Flores, 2008).
Finally, although this study examined two diverse host environments that are important in late blight (leaves and stored tubers), these environments do not represent all situations in which disease occurs. Tubers are highly dynamic organs in which starch and soluble sugars vary based on developmental stage, storage conditions and genotype (Coffin et al., 1987; Ross et al., 1994). Under some circumstances, free hexoses are less abundant than in the tubers tested here, and it would be interesting to assess how P. infestans adapts to these. Moreover, carbohydrates are distributed unequally in different portions of tubers, between sites within foliage and probably at different locations within a lesion (Daniele et al., 2003; Hajirezaei et al., 2003). How P. infestans responds to such differences and whether they affect its development are worthy topics for future study. The nutrient‐sensitive P. infestans promoters identified here could aid such analyses, along the lines of how an isocitrate lyase promoter–GFP fusion was employed to monitor the in planta metabolism of the wheat eyespot pathogen (Bowyer et al., 2000).
EXPERIMENTAL PROCEDURES
Sequence analysis
The P. infestans database at the Broad Institute was mined for genes of interest using keyword searches and BlastP queries. All gene models were evaluated manually. Models were corrected as needed using the Argo program (developed by R. Engels, Broad Institute, Cambridge, MA, USA).
Growth on laboratory media
Cultures were routinely maintained at 18 °C on 1.5% agar plates of rye–sucrose medium. This was obtained by germinating rye grain in water for 24 h, followed by blending, autoclaving for 30 min, removing major particulates using 1‐mm wire mesh and adding 2% sucrose. For gene or enzyme expression studies, 104 sporangia were spread on a 90‐mm polycarbonate membrane (0.4‐µm pore) laid on the surface of rye–sucrose agar, incubated for 5 days and harvested. Under such conditions, sporulation begins on the fifth day. To study the effect of sucrose, rye medium prepared as above but lacking sucrose was clarified by centrifugation, and the resulting broth was inoculated with 104/mL sporangia and incubated for 4 days. Half of the cultures were then amended with 2% sucrose, and tissue was harvested after 2 h. To study the effect of amino acids, cultures were grown for 3 days in defined media (Xu, 1982) with or without 1% casamino acids.
Infection of plant tissue
Inoculations were performed by dipping plant material into zoospore suspensions, which were obtained by placing sporangia in water (105/mL) for 2 h at 10 °C. The foliage of tomato plants (New Yorker) grown in soil for about 8 weeks was dipped into the zoospores, and incubated at 16 °C in a growth chamber with a 12‐h day/12‐h night cycle using Sylvania Grow‐Lux lamps at 154 W/m2. Leaflets were isolated for analyses at 0, 3 or 6 days post‐infection, and from mock‐infected material. Sporulation was typically first observed on the morning of the sixth day following inoculation.
Potato tuber infections were performed using cultivar Yukon Gold. These tubers were obtained from a commercial source approximately 8 weeks after harvest, and were stored at 10 °C for 1 week in the laboratory prior to analysis. This temperature is typical of most commercial warehouses. The tubers were cut into approximately 3.5‐mm slices, which were dipped into the zoospore suspensions and incubated at 16 °C in the dark. Sporulation began after 6 days, at which time infected and mock‐infected samples were harvested.
RNA analysis
Plant and hyphal tissues were frozen in liquid nitrogen and ground to a powder. In the case of tomato, this involved analysing the leaflets only, which were excised from the rest of the plant material. RNA was extracted using the Qiagen RNAEasy Plant Mini Kit (Valencia, CA, USA). The quality of RNA was assessed spectrophotometrically and by gel electrophoresis.
For qRT‐PCR, RNA was treated with DNAse and reverse‐transcribed with oligo‐dT using a first‐strand synthesis kit (Invitrogen, Carlsbad, CA, USA). Before initiating qRT‐PCR, standard PCR was performed to normalize cDNA concentrations between samples, using primers for the constitutive ribosomal protein S3a gene (Judelson et al., 2008). Based on these results, varying amounts of cDNA were used as templates for qRT‐PCR. This typically was 5 ng of cDNA from pure P. infestans samples and 50–100 ng from infected plants. qRT‐PCR was performed with hot‐start Taq polymerase in an iCycler (Bio‐Rad, Richmond, CA, USA), using the intercalation of SYBR Green as a reporter. Reactions involved one cycle of 95 °C for 8 min and 35 cycles of 95 °C for 20 s, 55 °C for 20 s and 72 °C for 30 s. Each biological treatment was performed with two replicates, with each analysed in duplicate. Relative expression was determined from the four resulting C T values using the ΔΔC T method and assuming a PCR efficiency of 1.9. Controls lacking reverse transcriptase were performed, and end‐point products were checked to confirm amplification of the desired bands. Primers usually targeted 175–225‐nucleotide regions near the 3′ end of each gene. However, when members of a gene family had similar sequences, it was sometimes necessary to target other regions to ensure specificity.
RNA blots were performed using 32P‐labeled probes (Kim and Judelson, 2003) using 1.5 µg of RNA from pure P. infestans samples and 15 µg from plant samples. Signal intensities were determined by phosphorimager analysis. After stripping probes, expression levels were normalised using a probe for elongation factor‐1α (EF‐1).
Enzyme assays
Hyphal cultures were disrupted using a Polytron homogenizer (Kinematica, Bohemia, NY, USA) in 50 mm sodium phosphate, pH 7.0. Clarified extracts were then assayed for enzyme activity, except for invertase for which the total extract was analysed to include particulate phase activity. Assays were performed using 50–250 µg protein at room temperature. Specific activity is expressed as micromoles of substrate converted per milligram of protein per minute, based on the linear portion of each reaction curve. Arginase was assayed by following the formation of urea from arginine using 1‐phenyl‐1,2‐propanedione‐2‐oxime (Corraliza et al., 1994). Hexokinase was measured using glucose as a substrate through a coupled reaction with glucose‐6‐phosphate dehydrogenase (Worthington, 1993). Histidine ammonia lyase was assayed by detecting the deamination of histidine to urocanate as described previously (Rechler, 1969), except that reactions were performed using 50 mm Tris‐Cl buffer at pH 7.5. Invertase was assayed using the 3,5‐dinitrosalicylic acid method to measure reducing sugars liberated from sucrose (Pan et al., 2005). Malate dehydrogenase was determined by following the conversion of oxaloacetate to malate, based on the oxidation of NADH (Worthington, 1993). Phosphoenolpyruvate carboxykinase was assayed by a carboxylation assay as described previously (Martín et al., 2007), except that reactions were performed in 50 mm Hepes buffer, pH 6.9.
Metabolite analysis
This was performed essentially as described previously (Johansen et al., 1996; Smith, 1969). Material was dried at 65 °C, ground in an electric mill and passed through a 40‐mesh wire screen. To measure free sugars (fructose, glucose, sucrose), samples were extracted using hot deionized water and analysed by high‐performance liquid chromatography (HPLC) with mass‐selective detection, using a Phenomenex Luna NH2 column with acetonitrile–water (78 : 22). To measure starch, samples were hydrolysed using amyloglucosidase, followed by HPLC determination of the glucose, with starch represented by total detected glucose minus free glucose multiplied by 0.9.
ACKNOWLEDGEMENTS
This work was supported by awards to H.S.J. by the National Research Initiative program of the United States Department of Agriculture‐Cooperative State Research, Education, and Extension Service.
REFERENCES
- Abood, J.K. and Losel, D.M. (2003) Changes in carbohydrate composition of cucumber leaves during the development of powdery mildew infection. Plant Pathol. 52, 256–265. [Google Scholar]
- Barelle, C.J. , Priest, C.L. , Maccallum, D.M. , Gow, N.A. , Odds, F.C. and Brown, A.J. (2006) Niche‐specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S. , Papadopoulos, M. , Schreiber, U. , Kaiser, W. and Roitsch, T. (2004) Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plant. 122, 419–428. [Google Scholar]
- Bialczyk, J. , Lechowski, Z. , Dziga, D. and Molenda, K. (2005) Carbohydrate and free amino acid contents in tomato plants grown in media with bicarbonate and nitrate or ammonium. Acta Physiol. Plant. 27, 523–529. [Google Scholar]
- Biemelt, S. and Sonnewald, U. (2006) Plant–microbe interactions to probe regulation of plant carbon metabolism. J. Plant Physiol. 163, 307–318. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. and Thomma, B.P.H.J. (2008) The complexity of nitrogen metabolism and nitrogen‐regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 72, 104–110. [Google Scholar]
- Bowyer, P. , Mueller, E. and Lucas, J. (2000) Use of an isocitrate lyase promoter‐GFP fusion to monitor carbon metabolism of the plant pathogen Tapesia yallundae during infection of wheat. Mol. Plant Pathol. 1, 253–262. [DOI] [PubMed] [Google Scholar]
- Brierley, E.R. , Bonner, P.L.R. and Cobb, A.H. (1996) Factors influencing the free amino acid content of potato (Solanum tuberosum) tubers during prolonged storage. J. Sci. Food Agric. 70, 515–525. [Google Scholar]
- Carpena‐Ruiz, R. , Sopena, A. and Ramon, A.M. (1989) Extraction of free amino acids from tomato leaves. Plant Soil, 119, 251–254. [Google Scholar]
- Coffin, R.H. , Yada, R.Y. , Parkin, K.L. , Grodzinski, B. and Stanley, D.W. (1987) Effect of low temperature storage on sugar concentrations and chip color of certain processing potato cultivars and selections. J. Food Sci. 52, 639–645. [Google Scholar]
- Corraliza, I.M. , Campo, M.L. , Soler, G. and Modolell, M. (1994) Determination of arginase activity in macrophages: a micromethod. J. Immunol. Methods, 174, 231–235. [DOI] [PubMed] [Google Scholar]
- Daniele, E. , Dommes, J. and Hausman, J.‐F. (2003) Carbohydrates and resistance to Phytophthora infestans in potato plants. Acta Physiol. Plant. 25, 171–178. [Google Scholar]
- Divon, H.H. and Fluhr, R. (2007) Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiol. Lett. 266, 65–74. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Wahl, R. , Horst, R.J. , Voll, L.M. , Usadel, B. , Poree, F. , Stitt, M. , Pons‐Kuhnemann, J. , Sonnewald, U. , Kahmann, R. and Kamper, J. (2008) Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis . Plant J. 56, 181–195. [DOI] [PubMed] [Google Scholar]
- Engstrom, K. and Stromberg, A. (1996) Changes in sugar content during induction of systemic acquired resistance to late blight caused by Phytophthora infestans (Mont) de Bary in potato. J. Phytopathol. 144, 33–36. [Google Scholar]
- Ferrer‐Lorente, R. , Fernandez‐Lopez, J.A. and Alemany, M. (2007) Estimation of the metabolizable energy equivalence of dietary proteins. Eur. J. Nutr. 46, 1–11. [DOI] [PubMed] [Google Scholar]
- Ferris, S.D. and Whitt, G.S. (1979) Evolution of the differential regulation of duplicate genes after polyploidization. J. Mol. Evol. 12, 267–317. [DOI] [PubMed] [Google Scholar]
- Gancedo, C. and Flores, C.L. (2008) Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72, 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo, J.M. (1998) Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62, 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirezaei, M.‐R. , Boernke, F. , Peisker, M. , Takahata, Y. , Lerchl, J. , Kirakosyan, A. and Sonnewald, U. (2003) Decreased sucrose content triggers starch breakdown and respiration in stored potato tubers (Solanum tuberosum). J. Exp. Bot. 54, 477–488. [DOI] [PubMed] [Google Scholar]
- Hohl, H.R. and Stoessel, P. (1976) Host–parasite interfaces in a resistant and a susceptible cultivar of Solanum tuberosum inoculated with Phytophthora infestans: tuber tissue. Can. J. Bot. 54, 900–912. [Google Scholar]
- Hohl, H.R. and Suter, E. (1976) Host–parasite interfaces in a resistant and a susceptible cultivar of Solanum tuberosum inoculated with Phytophthora infestans: leaf tissue. Can. J. Bot. 54, 1956–1970. [Google Scholar]
- Hu, G. , Cheng, P.Y. , Sham, A. , Perfect, J.R. and Kronstad, J.W. (2008) Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 69, 1456–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm, A. and Howlett, B.J. (2001) Pathogenicity genes of phytopathogenic fungi. Mol. Plant Pathol. 2, 241–255. [DOI] [PubMed] [Google Scholar]
- Johansen, H.N. , Glitso, V. and Knudsen, K.E.B. (1996) Influence of extraction solvent and temperature on the quantitative determination of oligosaccharides from plant materials by high‐performance liquid chromatography. J. Agric. Food Chem. 44, 1470–1474. [Google Scholar]
- Judelson, H.S. , Ah‐Fong, A.M. , Aux, G. , Avrova, A.O. , Bruce, C. , Cakir, C. , da Cunha, L. , Grenville‐Briggs, L. , Latijnhouwers, M. , Ligterink, W. , Meijer, H.J. , Roberts, S. , Thurber, C.S. , Whisson, S.C. , Birch, P.R. , Govers, F. , Kamoun, S. , Van West, P. and Windass, J. (2008) Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol. Plant–Microbe Interact. 21, 433–447. [DOI] [PubMed] [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. , Beale, M.H. , Hammond‐Kosack, K. and Rudd, J.J. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant–Microbe Interact. 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. and Judelson, H.S. (2003) Sporangia‐specific gene expression in the oomyceteous phytopathogen Phytophthora infestans . Eukaryot. Cell, 2, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebherz, H.G. and Rutter, W.J. (1969) Distribution of fructose diphosphate aldolase variants in biological systems. Biochemistry, 8, 109–121. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.C. , Pan, X. , Harashima, T. , Cardenas, M.E. , Xue, Y. , Hirsch, J.P. and Heitman, J. (2000) The G protein‐coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae . Genetics, 154, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, J.S. , Ashton, A.R. , Govers, F. and Hardham, A.R. (2001) Isolation and characterization of four genes encoding pyruvate, phosphate dikinase in the oomycete plant pathogen Phytophthora cinnamomi . Curr. Genet. 40, 73–81. [DOI] [PubMed] [Google Scholar]
- Martín, M. , Plaxton, W.C. and Podésta, F.E. (2007) Activity and concentration of non‐proteolyzed phosphoenolpyruvate carboxykinase in the endosperm of germinating castor oil seeds: effects of anoxia on its activity. Physiol. Plant. 130, 484–494. [Google Scholar]
- Mendgen, K. and Hahn, M. (2002) Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 7, 352–356. [DOI] [PubMed] [Google Scholar]
- Moore, P.A. , Sagliocco, F.A. , Wood, R.M. and Brown, A.J. (1991) Yeast glycolytic mRNAs are differentially regulated. Mol. Cell. Biol. 11, 5330–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K. , Kunihiro, S. , Sano, M. , Zhang, Y. , Eto, S. , Chang, Y.C. , Suzuki, T. , Jigami, Y. and Machida, M. (2000) Comprehensive cloning and expression analysis of glycolytic genes from the filamentous fungus, Aspergillus oryzae . Curr. Genet. 37, 322–327. [DOI] [PubMed] [Google Scholar]
- Pan, Q.‐H. , Li, M.‐J. , Peng, C.‐C. , Zhang, N. , Zou, X. , Zou, K.‐Q. , Wang, X.‐L. , Yu, X.‐C. , Wang, X.‐F. and Zhang, D.‐P. (2005) Abscisic acid activates acid invertases in developing grape berry. Physiol. Plant. 125, 157–170. [Google Scholar]
- Rechler, M.M. (1969) The purification and characterization of L‐histidine ammonia‐lyase (Pseudomonas). J. Biol. Chem. 244, 551–559. [PubMed] [Google Scholar]
- Roitsch, T. , Balibrea, M.E. , Hofmann, M. , Proels, R. and Sinha, A.K. (2003) Extracellular invertase: key metabolic enzyme and PR protein. J. Exp. Bot. 54, 513–524. [DOI] [PubMed] [Google Scholar]
- Ross, H.A. , Davies, H.V. , Burch, L.R. , Viola, R. and McRae, D. (1994) Developmental changes in carbohydrate content and sucrose degrading enzymes in tuberising stolons of potato (Solanum tuberosum). Physiol. Plant. 90, 748–756. [Google Scholar]
- Sheard, J. and Farrar, J.F. (1987) Transport of sugar in Phytophthora palmivora Butl. New Phytol. 105, 265–272. [Google Scholar]
- Slamovits, C.H. and Keeling, P.J. (2006) Pyruvate‐phosphate dikinase of Oxymonads and Parabasalia and the evolution of pyrophosphate‐dependent glycolysis in anaerobic eukaryotes. Eukaryot. Cell 5, 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. (1969) Removing and analyzing total nonstructural carbohydrates from plant tissue. Wis. Agric. Exp. Sta. Res. Rep. No. 41. [Google Scholar]
- Solomon, P.S. , Tan, K.‐C. and Oliver, R.P. (2003) The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 4, 203–210. [DOI] [PubMed] [Google Scholar]
- Unkles, S.E. , Logsdon, J.M. , Robison, K. , Kinghorn, J.R. and Duncan, J.M. (1997) The tigA gene is a transcriptional fusion of glycolytic genes encoding triose‐phosphate isomerase and glyceraldehyde‐3‐phosphate dehydrogenase in oomycota. J. Bacteriol. 179, 6816–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. , Shen, W. , Dauk, M. , Wang, F. , Selvaraj, G. and Zou, J. (2004) Targeted gene disruption of glycerol‐3‐phosphate dehydrogenase in Colletotrichum gloeosporioides reveals evidence that glycerol is a significant transferred nutrient from host plant to fungal pathogen. J. Biol. Chem. 279, 429–435. [DOI] [PubMed] [Google Scholar]
- Wong, K.H. , Hynes, M.J. and Davis, M.A. (2008) Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell 7, 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington, V. (1993) Worthington Enzyme Manual. Lakewood, NJ: Worthington Biochemical Corporation. [Google Scholar]
- Xu, R. (1982) A defined media for Phytophthora . Acta Mycol. Sin. 1, 40–47. [Google Scholar]
- Yan, H.Z. and Liou, R.F. (2006) Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genet. Biol. 43, 430–438. [DOI] [PubMed] [Google Scholar]


