Figure 2.
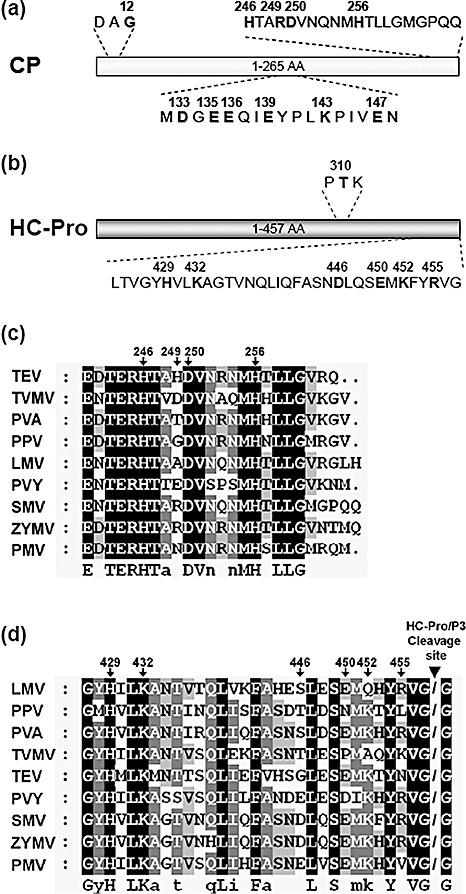
Identification of amino acid residues crucial for the coat protein–helper component‐proteinase (CP–HC‐Pro) interaction in a yeast two‐hybrid system (YTHS). Schematic representations of Soybean mosaic virus (SMV) CP (a) and HC‐Pro (b), showing the amino acid positions of the site‐directed mutations tested in this study. Alignment of the C‐terminal amino acid sequences of potyviral CPs (c) and HC‐Pros (d). Sequences were aligned by ClustalX2. Arrows indicate the amino acid positions on SMV CP and HC‐Pro. Arrowhead indicates the location of the HC‐Pro self‐cleavage site. Potyviral sequence data were obtained from the National Center for Biotechnology Information (NCBI) database: TEV, Tobacco etch virus (NC001555); TVMV, Tobacco vein mottling virus (NC001768); PVA, Potato virus A (NC004039); PPV, Plum pox virus (NC001445); LMV, Lettuce mosaic virus (NC003605); PVY, Potato virus Y (NC001616); SMV, Soybean mosaic virus (FJ807700); ZYMV, Zucchini yellow mosaic virus (NC003224); PMV, Peanut mottle virus (NC002600).
