Figure 3.
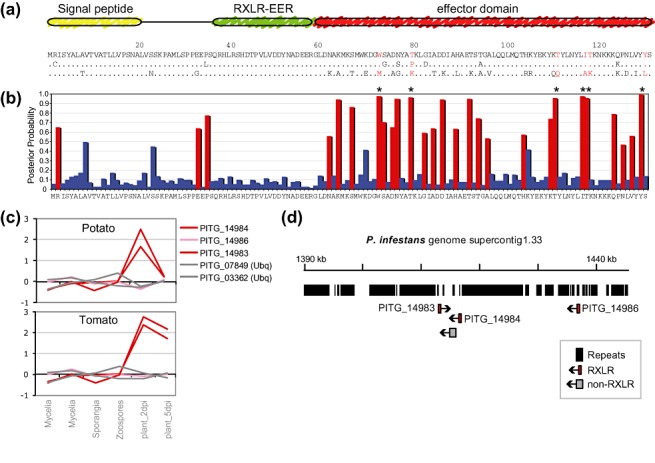
RXLR effector genes typically show adaptive selection in their C‐termini, are induced in plants and occur in clusters in the genome. The figure depicts the features of a representative RXLR gene cluster of Phytophthora infestans (Haas et al., 2009). (a) Domain structure and sequence variability of three paralogues of RXLR family 6 of P. infestans (PITG_14983, PITG_14984 and PITG_14986, top to bottom; based on Haas et al., 2009). Residues with evidence of positive selection (b) are highlighted in red. Dots in the alignment represent identical amino acid residues. (b) Positive selection analyses based on the methods described in Win et al. (2007). Posterior probabilities (blue, red) for the site class with expected ω value >1 (ω= 21.07706) and P= 0.16379 estimated under the model M8 in the paml program (http://abacus.gene.ucl.ac.uk/software/paml.html). Positively selected sites are shown in red. Asterisks label residues with P > 95%. (c) Relative oligonucleotide microarray expression levels at different developmental stages and during infection of potato and tomato plants 2 and 5 days post‐inoculation (dpi). Two RXLR genes are induced in plants (red lines) and one is not (pink line). Two constitutive ubiquitin genes (Ubq) are shown as controls (grey lines) (see Haas et al., 2009 for experimental details). (d) Genome browser view of ∼55‐kbp region of the P. infestans genome (supercontig 1.33) containing the cluster of related RXLR genes. The high content of repetitive sequences is evidenced by the black bars (repeats).
