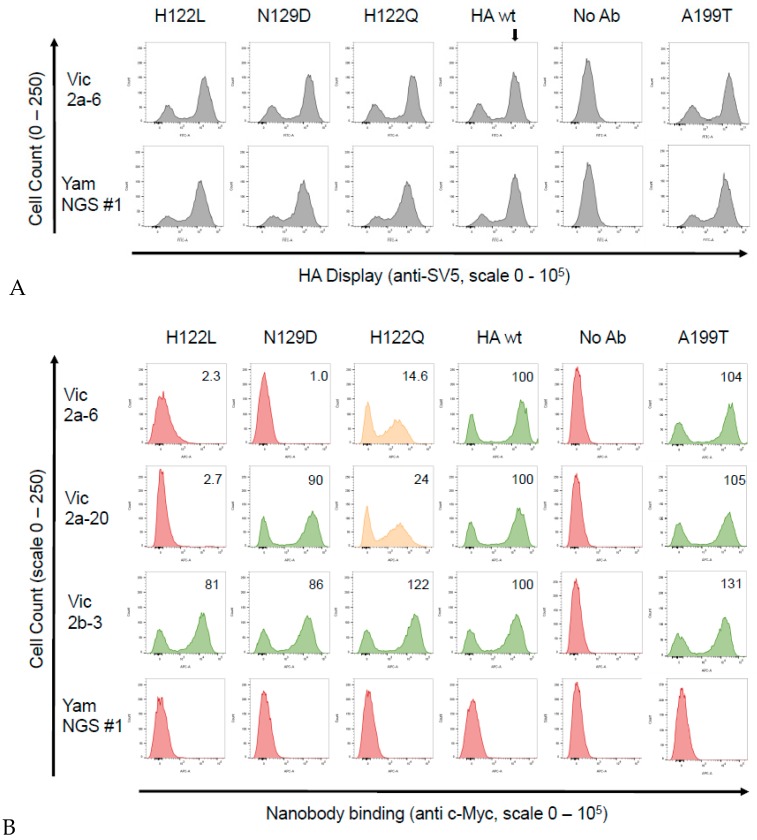
Figure 4.
Binding of the sdAbs to the B-Victoria single residue mutant HA0’s. Candidate single residue mutations predicted to correlate with lineage-specific binding and a naturally occurring substitution H122Q, which differentiated between the B-Victoria and B-Yamagata strains, were generated and binding was tested using yeast display and flow cytometry. (A) Detection of the HA0 display level, using detection of the SV5 epitope tag attached to the HA0, showing that the mutations had no effect on the HA0 display level relative to the wild-type (wt) HA0 (indicated by vertical arrow). (B) Flow cytometry histograms showing binding of the Vic2a-6, Vic2a-20, Vic2b-3 and the YamNGS#1 to the wild-type HA (B/Brisbane/60/2008 precursor HA0) and selected mutants (the positive population is the right peak, whereas, the left peak shows the unbound and unstained populations). Mutations that completely eliminated sdAb binding are shown in red, those that partially affected binding are shown in orange and those that had no effect are shown in green. We determined the extent of sdAb binding by dividing the geometric mean fluorescence intensity (MFI) of each sdAb mutant pair by the value of the MFI of the wild-type B-Vic HA sdAb interaction and the resulting ratio, normalised to a percentage value of the wild-type interaction. Relative binding of the sdAbs was categorised as ‘no binding’ (in red)—< 5%; ‘intermediate binding’ (in orange)—between 20–40%; and ‘strong binding’ (in green)—> 40%; shown in the upper right-hand quadrant of the flow cytometry histograms. Graphs shown are a representative of at least two independent experiments.