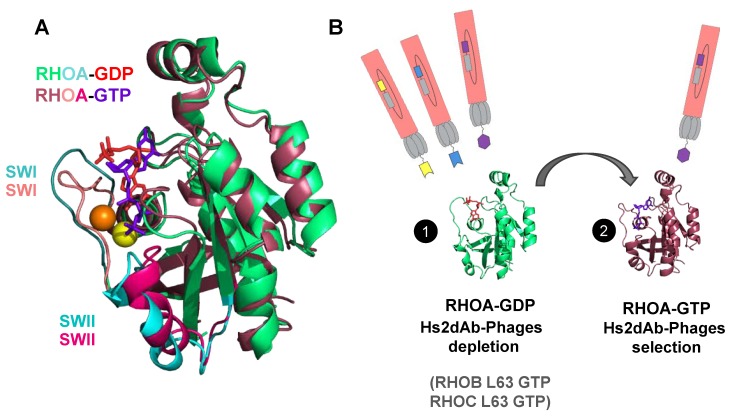
Figure 1.
Antibody phage display selection of GTP-bound RHO conformational nanobodies. (A) View of the structure of RHOA-GTP V14 mutant (shown in wine-red and pink, Protein Data Base (PDB): 1a2b) superimposed with the structure of RHOA-GDP (shown in green and cyan, PDB: 1ftn). RHOA G14V mutant in the active state is bound to the GTP (purple-blue nucleotide) and Mg2+ (shown as a yellow sphere). RHOA displays the inactive conformation bound to GDP (red nucleotide) and Mg2+ (shown as an orange sphere). The structural alignment in this view shows a subtle closure of the switch I and switch II loops (SWI and SWII loops currently in cyan in RHOA-GDP and in pink in RHOA-GTP) around the phosphate gamma and the Mg2+ (orange to yellow). (B) Scheme of the subtractive phage display enrichment of hs2dAb to GTP-bound RHOA (wine-red) by depletion with the inactive GDP-bound state (green) and with the GTP-bound state of RHOB and RHOC (see Methods). The phages presenting hs2dAb are shown in pink/grey. The RHOA structures were produced using PyMOL software 2.1 (Schrödinger, Mannheim, Germany).