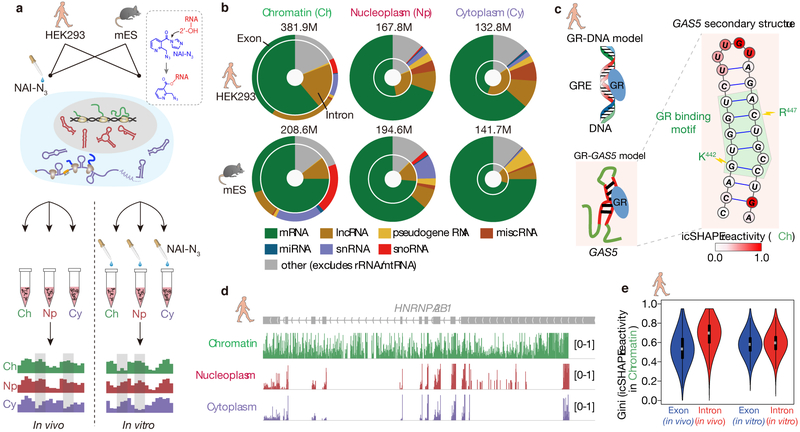
Fig 1 ∣. Chromatin fractions are enriched for pre-mRNA and lncRNA structures.
a, Experimental overview of the icSHAPE protocol. The dashed box highlights the chemical structure of NAI-N3 and its covalent bond with the 2'-OH group of RNA, which allows probing of RNA structures inside living cells. b, Donut charts showing read distributions of different RNA types in the three cellular compartments. The outer circles represent exon coverage while the inner circles represent intron coverage. c, GAS5 RNA secondary structure with icSHAPE reactivity scores shown in color. The nucleotides outlined in red interact with GR amino acids, shown in blue. d, UCSC tracks showing icSHAPE reactivity scores (y-axis), along the RNA sequence. 1 denotes unstructured (single-stranded) regions, and 0 denotes fully-structured regions. e, Violin plot of Gini index of icSHAPE data in exon versus in intron. The thick black bar in the center of the Violin plot represents the interquartile range, the thin black line extended from it represents the 95% confidence intervals, and the white dot is the median. The numbers of sliding windows (width = 20nt) on the respective regions from the left to the right are n=18930, n=5926, n=51409, n=82648.