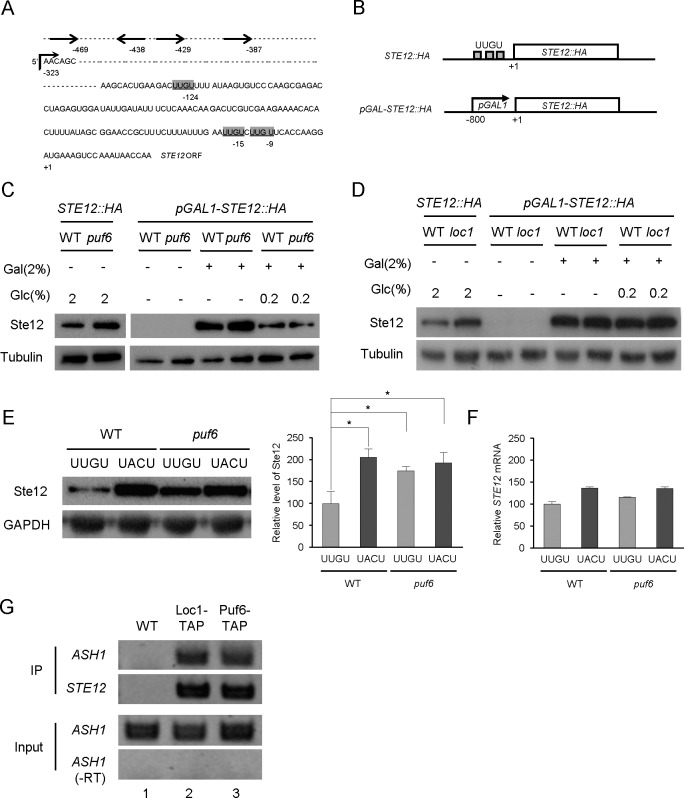
Fig 3. 5’UTR-dependent repression of the STE12 mRNA.
(A) Schematic representation of the UUGU sequences in the 5’-UTR of STE12 mRNA. The AUG start codon is indicated as +1 in the STE12 ORF. UUGU sequences were found at the -124, -15, and -9 positions. The 5’-UTR starts at -323 position as indicated with a right angle arrow. Sequences indicated with arrows represent the pheromone responsive elements (PREs). (B) Diagrams showing the wild-type STE12::HA and pGAL-STE12::HA constructs. The pGAL-STE12::HA plasmid carried a 800-bp fragment containing the GAL1 promoter region. (C) Western blot analysis of Ste12-HA proteins prepared from wild-type and loc1 cells carrying the STE12::HA or pGAL-STE12::HA plasmids. For STE12::HA (lanes 1 and 2), cells grown to the early log phase (OD600 = 1.0) in 2% glucose medium were used as a control. For galactose induction, cells were grown in SC medium containing 2% raffinose to the early log phase (OD600 = 1.0), and induced with 2% galactose or 2% galactose plus 0.2% glucose for 2 hours. Lanes 3 and 4, 2% raffinose as a sole carbon source; lanes 5 and 6, 2% galactose was added for GAL1 promoter induction; lanes 7 and 8, 2% galactose plus 0.2% glucose. Tubulin was detected as a loading control. (D) Western blot analysis of Ste12-HA proteins prepared from wild-type and puf6 cells carrying STE12::HA or pGAL-STE12::HA plasmids. Cultures and protein analysis were performed essentially as described in (C). (E) Ste12-HA protein levels in cells carrying the wild-type UUGU-STE12-HA or mutant UACU-STE12-HA construct, as revealed by Western blot analysis. Graphs represent quantification of Ste12-HA to GAPDH ratio (n = 2 independent replicates). Values are mean ± SD. *p < 0.05. (F) RNA prepared from the cultures listed in (E) was analyzed by quantitative RT-PCR. STE12 mRNA expression was normalized against ACT1 mRNA expression (error bars, mean + S.D.). (G) RNA immunoprecipitation of Loc1-TAP and Puf6-TAP from the cell extracts of the wild-type or TAP-tagged strain, followed by RNA purification and RT-PCR. STE12-specific primers were used for PCR. ASH1 mRNAs were detected as a binding control.–RT indicates RT-PCR without reverse transcriptase.