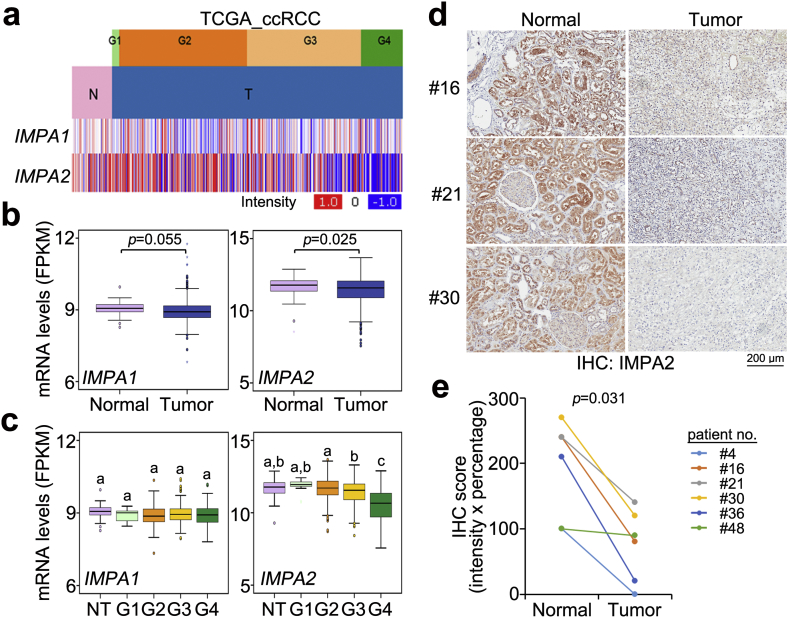
Fig. 1.
IMPA2 downregulation significantly correlates with tumorigenesis and cancer progression in ccRCC. (a) Heatmap for the transcriptional profiles of the IMPA1 and IMPA2 genes in normal (N) tissues and primary tumors (T) of different grades from the TCGA ccRCC database. (b and c) Boxplot for the mRNA levels of IMPA1 and IMPA2 in normal tissues (n = 72) and primary tumors (n = 534) (b) or normal tissues (NT, n = 72) and tumors with grade G1 (n = 14), G2 (n = 229), G3 (n = 207) or G4 (n = 76) (c), as shown in a. In b and c, the band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. The significant differences were analyzed by independent sample t-test and Tukey's post-hoc test, respectively. Different letters in the boxplot indicate the statistical significance at p < .05. (d) Immunohistochemistry (IHC) staining for the IMPA2 protein in paired normal and tumor tissues from ccRCC patients #16, #21 and #30. (e) IHC intensity of the IMPA2 protein in paired normal and tumor tissues derived from 6 ccRCC patients. Statistical significance was estimated with a non-parametric Wilcoxon signed rank test.