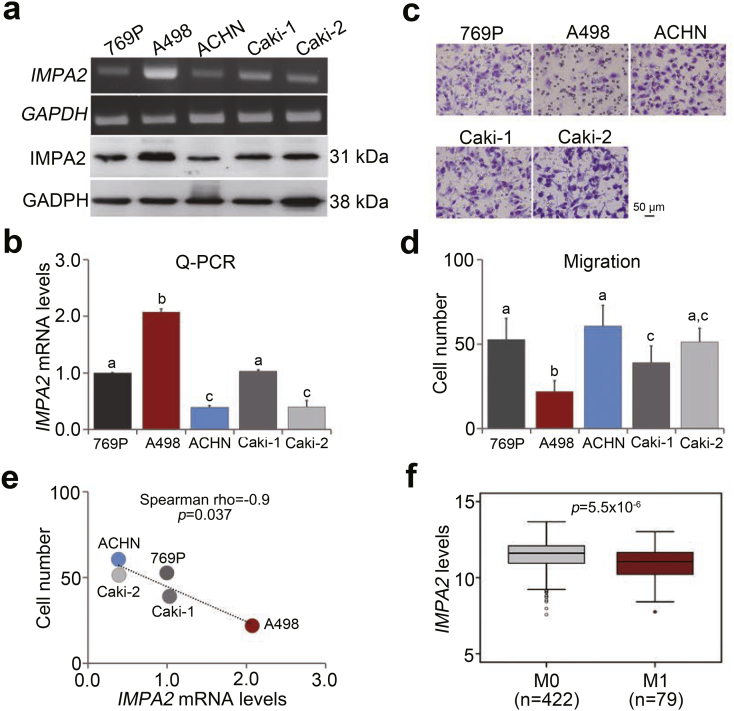
Fig. 3.
IMPA2 expression is inversely associated with metastatic potential in ccRCC cells. (a and b) IMPA2 levels determined by RT-PCR (a, upper), Western blotting (a, bottom) and quantitative PCR (b) in the tested ccRCC cell lines 769P, A498, ACHN, Caki-1 and Caki-2. (c and d) Giemsa staining (c) and data from three independent experiments (d) for the migrated cells in the 3-hour transwell assay for the ccRCC cells. In b and d, error bars represent the mean ± SEM of data obtained from three independent experiments and the statistical differences were analyzed by Kruskal Wallis test. Different letters in each column of b and d indicate the statistical significance at p < .05. (e) Scatchard plot for the endogenous IMPA2 mRNA levels from b and the migrated cell number from d. Spearman's correlation test was used to determine the statistical significance. (f) Transcriptional profile of the IMPA2 gene in primary tumors from TCGA ccRCC patients without (M0) or with (M1) metastatic progression. The band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. The independent sample t-test was used to estimate the statistical significance.