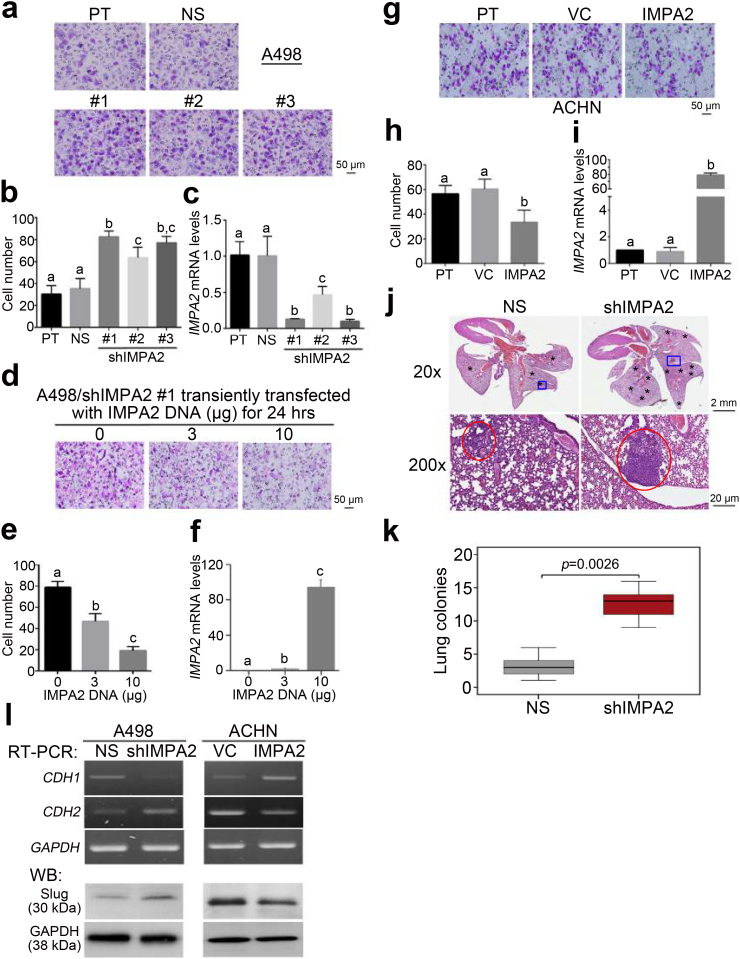
Fig. 4.
IMPA2 downregulation promotes metastatic progression in ccRCC cells. (a–c) Giemsa staining (a) and cell number (b) for the migrated cells in 3-hour transwell assays and quantitative PCR for IMPA2 gene expression (c) in A498 cells transfected without (parental, PT) or with a nonsilencing (NS) sequence or one of 3 independent IMPA2 shRNA clones. (d-f) Giemsa staining (d) and cell number (e) for the migrated cells in 3-hour transwell assays and quantitative PCR for IMPA2 gene expression (f) in IMPA2 shRNA clone #1 (targeting 3’ UTR)-overexpressing A498 cells transiently transfected with exogenous IMPA2 DNA in the designated amounts for 24 h. (g-i) Giemsa staining (g) and cell number (h) for the migrated cells in 3-hour transwell assays and quantitative PCR for IMPA2 gene expression (i) in ACHN cells stably transfected without (parental, PT) or with a vector control (VC) or a vector containing the IMPA2 gene. In b, c, e, f, h, and i, error bars represent the mean ± SEM of data obtained from three independent experiments. The different letters in each column of b, c, e, f, h, and i indicate the statistical significance at p < .05 as determined by Friedman test. (j) Hematoxylin/eosin staining for lung tissues derived from mice transplanted with A498 cells transfected with nonsilencing (NS) sequences or IMPA2 shRNA via tail-vein injections. The symbols (*) and red circles indicate the colonies of A498 cells. (k) Boxplot for the lung colonies counted from tumor-bearing mice (n = 5) as shown in j. The band inside the box is the second quartile (the median). The upper and lower lines of box are the third and first quartiles, respectively. Box plots have lines extending vertically from the whiskers indicating minimum and maximum of all of the data. The individual points indicate outliers. Mann-Whitney U test was used to estimate the statistical significance. (l) RT-PCR for CDH1, CDH2 and GAPDH genes and Western blotting (WB) for Slug and GAPDH proteins derived from A498 cells without (NS) or with IMPA2 knockdown (shIMPA2) and ACHN cells without (VC) or with IMPA2 overexpression (IMPA2). GAPDH was used as internal controls for RT-PCR and Western blot analyses. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)