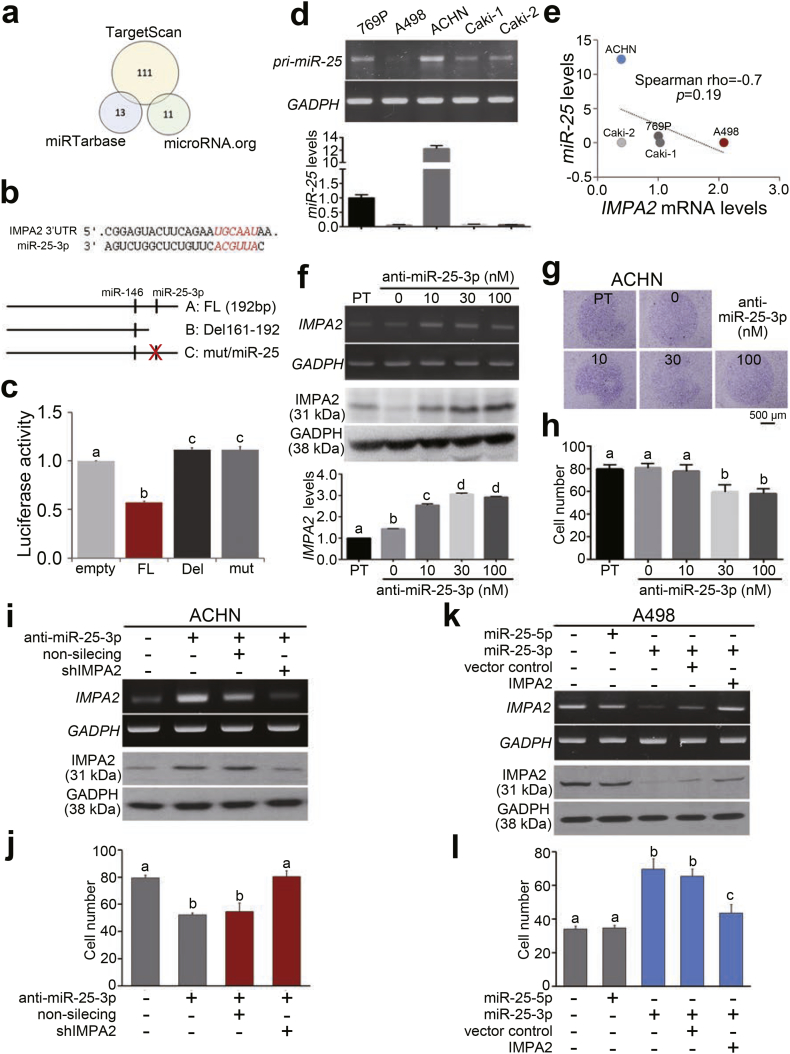
Fig. 5.
IMPA2 downregulation is mediated by the miR-25 posttranscriptional inhibition in ccRCC cells. (a) An in silico analysis of IMPA2-targeting microRNAs using the TargetScan, miRTarbase and microRNA.org databases. The numbers indicate the microRNAs that are computationally predicted to bind the 3′ untranslated region (3′ UTR) of the IMPA2 transcript. (b) Experimental designs for the cloning of the IMPA2 3′ UTR with no alteration (FL, full-length), with a deletion (Del) among base pairs (bp) 161–192 (numbered from the bp next to the stop codon) or with a mutation (mut) at the miR-25 binding sequences into the luciferase-expressing pmirGLO plasmid. (c) Luciferase activity in ACHN cells transiently transfected with the empty pmirGLO plasmid or the plasmid containing the IMPA2 3’ UTR without alterations (FL) or with a deletion (Del 161–192) or miR-25 binding site mutation. One-way ANOVA with Tukey's test was used to estimate the statistical significance. (d) RT-PCR (upper) and quantitative PCR (bottom) analyses for miR-25 and GAPDH expression in a panel of ccRCC cell lines. GAPDH was used as an internal control and was utilized to normalize the miR-25 levels in quantitative PCR. (e) Scatchard plot for the endogenous IMPA2 mRNA levels from Fig. 3b and the miR-25 levels from d. Spearman's correlation test was used to determine the statistical significance. (f) RT-PCR (upper), Western blot (middle) and quantitative PCR (Q-PCR, bottom) analyses for IMPA2 and GAPDH expression in ACHN cells transiently transfected with anti-miR-25-3p oligonucleotides at the designated concentrations for 24 h. GAPDH was used as an internal control. (g and h) Giemsa staining (g) and cell number (h) for the migrated cells in 3-h transwell assays for ACHN cells transiently transfected with anti-miR-25 oligonucleotides at the designated concentrations for 24 h. (i) RT-PCR (upper) and Western blot (bottom) analyses for IMPA2 and GAPDH expression in ACHN cells transiently transfected without or with anti-miR-25-3p oligonucleotides combining with non-silencing or IMPA2 shRNA for 24 h. GAPDH was used as an internal control. (j) Migrated cell number of ACHN cells without or with the designed transfections as shown in i. (k) RT-PCR (upper) and Western blot (bottom) analyses for IMPA2 and GAPDH expression in A498 cells without or with the overexpression of miR-25-5p or miR-25-3p combining without (vector control) or with the restoration of exogenous IMPA2 gene. GAPDH was used as an internal control. (l) Migrated cell number of A498 cells without or with the designed transfections as shown in k. In c, f, h, j and l, error bars represent the mean ± SEM of data obtained from three independent experiments. The different letters in each column of c, f, h, j and l indicate the statistical significance at p < .05 as determined by Friedman test.