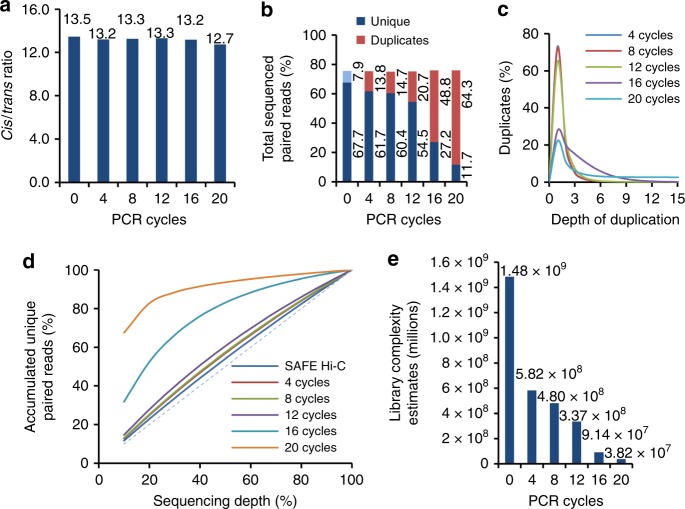
Fig. 2.
Amplification increases PCR duplicates and reduces Hi-C library complexity of the Drosophila genome. a Cis paired reads were uniquely mapped on the same chromosome, and trans paired reads were mapped on different chromosomes. SAFE (simplified, amplification-free, and economically efficient process) Hi-C is referred to as 0 PCR cycle in figures. b Percentage of unique paired reads and duplicates that can be aligned in the total sequenced paired reads. The light blue bar shows the duplicates in SAFE Hi-C libraries, which we kept as unique as no amplification was involved in the library preparation process. c Percentage of ligates duplicated at different depths introduced by PCR amplification. d Accumulated percentage of unique paired reads against the percentage of sequencing depth. e Library complexity estimates. For SAFE Hi-C, the complexity was estimated as described in Methods