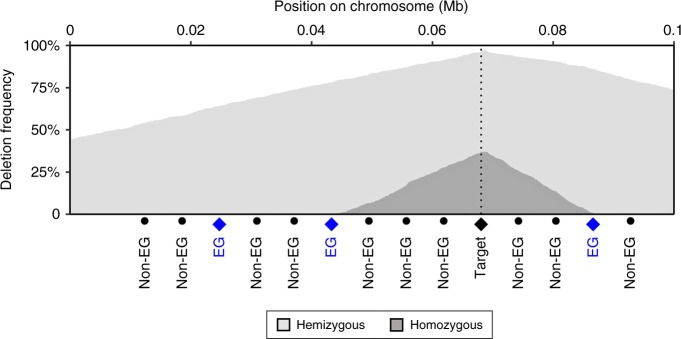
Fig. 1.
Linear deletion limitation illustrated by simulation. Hypothetically, the frequency of homozygous deletions in a recurrently deleted region should approach to zero at the nearest essential gene (EG). To illustrate this phenomenon, we simulated frequency of hemizygous and homozygous deletions across a fictive chromosomal region, harboring a deletion target gene surrounded by essential and non-essential genes. In these simulations, we required both copies of the target gene to be inactivated, however potentially through a variety of different mechanisms: regional deletion, whole-arm deletions, point mutation, or reduplication of the mutated chromosomes accompanied by deletion of the normal chromosome. The probability of each type of inactivation was determined by fixed probabilities. This plot shows the results across 1000 simulated chromosome pairs, and probabilities 20% for regional deletion, 7% for point mutation, and 1% for reduplication. Similar results were obtained with other parameter values. Regardless of parameter values, the frequency of homozygous deletions drops to zero at the essential genes located closer to the target gene