Figure 5.
Intestinal Th17 Cells Elicited by SFB and C. rodentium Have Distinct Transcriptional Profiles
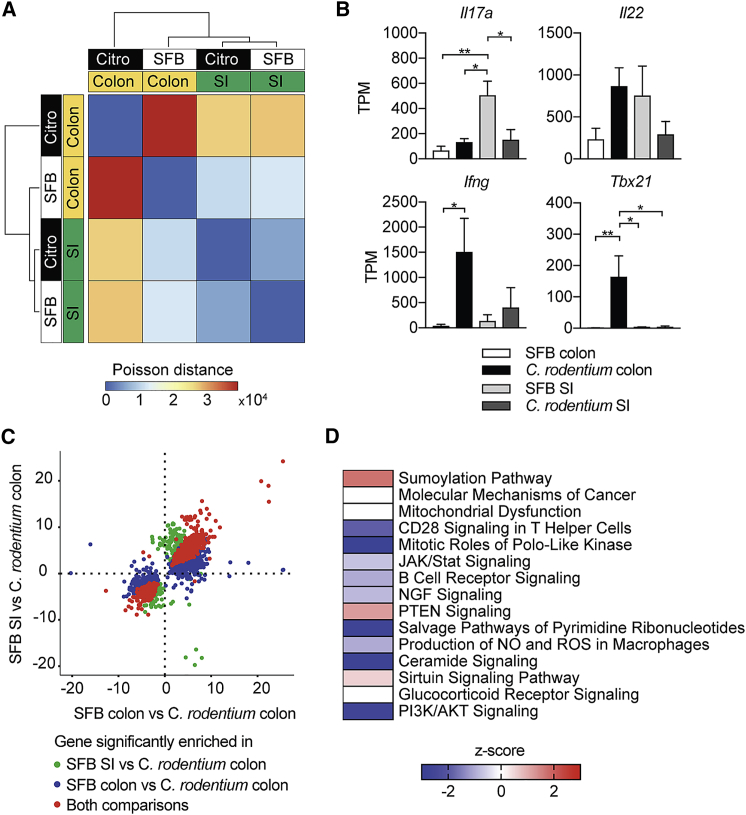
(A) RNA-sequencing analysis of Th17 cells from the SI and colon of C. rodentium (Citro)-infected and SFB-colonized mice, presented as heatmap of condition similarity showing Poisson distance between experimental groups.
(B) Gene expression in Th17 cells from the SI and colon of C.-rodentium-infected and SFB-colonized mice, presented as normalized read counts (TPM).
(C) Scatter plot of differentially expressed genes (log2-fold change) in SFB SI versus C. rodentium colon against SFB colon versus C. rodentium colon. Each dot represents a gene differentially expressed (adjusted p < 0.05) in at least one comparison. Specifically, genes in green are significantly different in the SFB SI versus C. rodentium colon comparison; genes in blue are significantly different in SFB colon versus C. rodentium colon comparison; and genes in red are significantly different in both comparisons.
(D) Top 15 canonical pathways in colonic SFB- versus C. rodentium-induced Th17 cells identified by ingenuity pathway analysis (IPA) and ranked on p value. The colors indicate activation Z-score of each pathway with red corresponding to positive Z-scores, blue to negative Z-scores, and white to no activity pathway available. ∗p < 0.05, ∗∗p < 0.01 by one-way ANOVA with Tukey’s post-test. See also Figure S5.