Figure 1.
Ectopic Recruitment of NDP52 to Mitochondria Can Initiate Mitophagy by Recruiting ATG Machinery
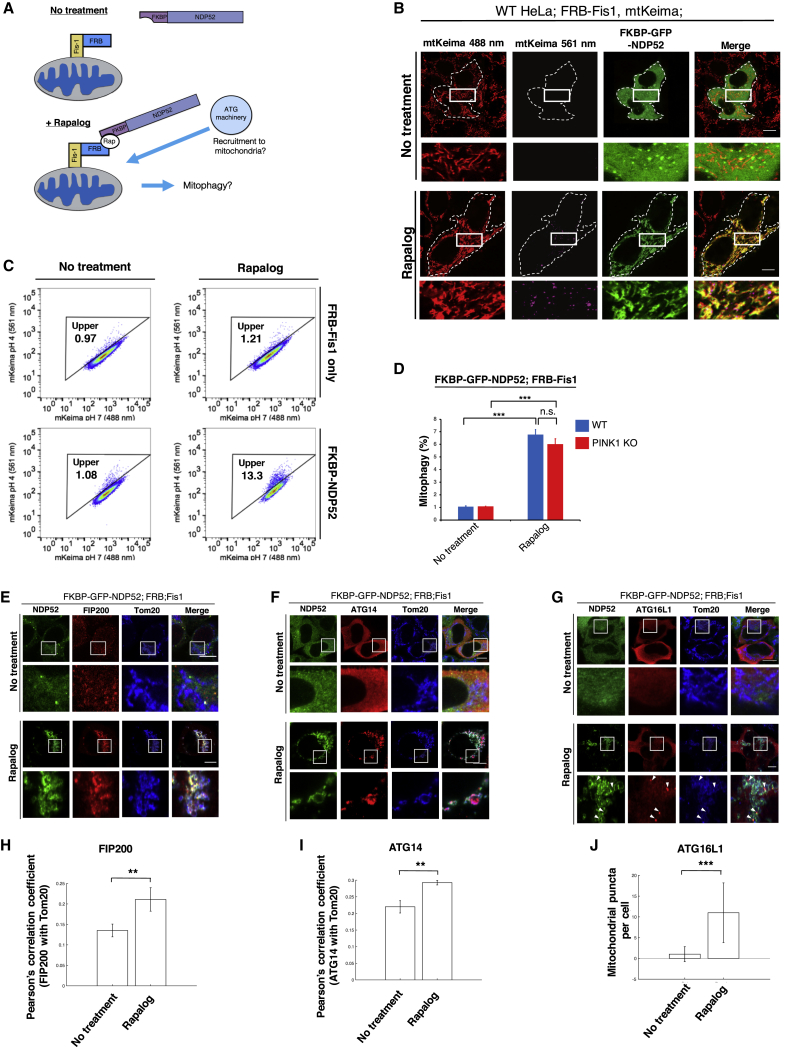
(A) Schematic of chemically inducible dimerization assay to control mitochondrial localization of NDP52.
(B) Live confocal imaging of HeLa cells stably expressing mito-mKeima and FRB-Fis1 and transiently expressing FKBP-GFP-NDP52. Cells were treated with Rapalog for 24 h. Cytosolic mitochondria (488 nm; red) and mitolysosomes (561 nm; purple) are shown.
(C) FACS plots showing mito-mKeima ratio (561/488 nm). HeLa cells stably expressing mito-mKeima, FRB-Fis1, and FKBP-GFP-NDP52 were treated with Rapalog for 24 h.
(D) Quantification of mito-mKeima ratiometric FACS analysis of WT or PINK1 KO HeLa cells stably expressing mito-mKeima, FRB-Fis1, and FKBP-GFP-NDP52 after 24 h of Rapalog treatment. n = 3 biological replicates.
(E–G) Confocal images of HeLa cells stably expressing mito-mKeima, FRB-Fis1, and FKBP-GFP-NDP52. Cells were transfected with (E) HA-FIP200, (F) HA-ATG14, and (G) Flag-ATG16L1. Cells were then treated with Rapalog for 24 h, fixed, then immunostained for Tom20 with either HA or Flag.
(H) Quantification of FIP200 recruitment to mitochondria by Pearson’s correlation coefficient as in (E).
(I) Quantification of ATG14 recruitment to mitochondria by Pearson’s correlation coefficient as in (F).
(J) Quantification of the number of ATG16L1 foci on mitochondria before and after Rapalog treatment as in (G). Arrows indicate foci of ATG16L1 colocalized with Tom20.
All FACS quantifications: n = 3. Data are represented as mean ± SD. p value: ∗ = < 0.05; ∗∗ < 0.01; ∗∗∗ < 0.001; ∗∗∗∗ < 0.00001; ns., not significant. Scale bars: 10 μm.
See also Figure S1.