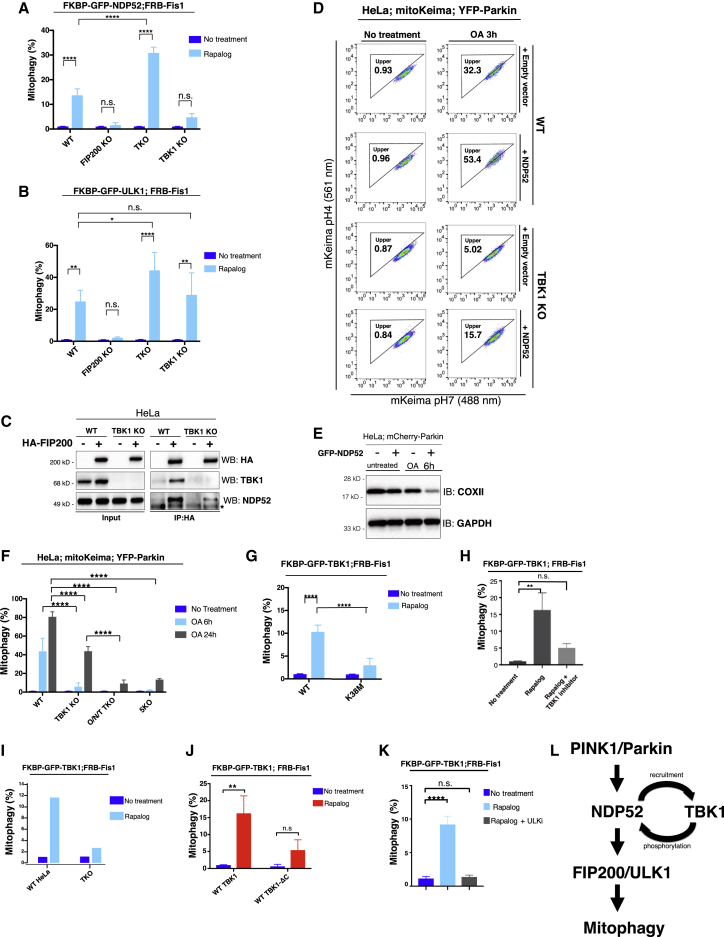
Figure 6.
TBK1 Mediates NDP52-ULK1 Complex Mitophagy
(A) Quantification of FACS ratiometric analysis of mito-mKeima. WT, FIP200 KO, TKO (OPTN, NDP52 Tax1bp1 triple KO), or TBK1 KO cells stably expressing FKBP-GFP-NDP52, FRB-Fis1, and mito-mKeima treated with Rapalog for 24 h.
(B) Quantification of FACS analysis of mito-mKeima. WT and KO cells as in (A), stably expressing FKBP-GFP-ULK1, FRB-Fis1, and mito-mKeima treated with 24 h Rapalog.
(C) Western analysis of HA-FIP200 immunoprecipitation in WT or TBK1 KO HeLa cells. Endogenous NDP52 and TBK1 are immunoblotted.
(D) FACS plots of mito-mKeima ratiometric analysis. WT or TBK1 KO cells stably expressing mito-mKeima and YFP-Parkin, and transiently expressing BFP-NDP52 were treated with OA for 3 h.
(E) Western analysis. Cells stably expressing mCherry-Parkin with or without GFP-NDP52 were treated with OA for 6 h.
(F) Quantification of FACS ratiometric analysis of mito-mKeima. WT, FIP200 KO, TKO, or TBK1 KO cells, all stably expressing mito-mKeima and YFP-Parkin, were treated with OA for 6 or 24 h.
(G) Quantification of FACS ratiometric analysis of mito-mKeima. Cells stably expressing FRB-Fis1 and either WT FKBP-GFP-TBK1 or kinase-dead FKBP-GFP-TBK1 K38M were treated with Rapalog for 24 h.
(H) mito-mKeima FACS analysis of cells stably expressing FRB-Fis1 and FKBP-TBK1 after mitochondrial localization of TBK1 with Rapalog with or without ULK1/2 inhibitor, MRT68921 at 1 μM.
(I) Quantification of FACS ratiometric analysis of mito-mKeima of WT or TKO stably expressing FRB-Fis1 and WT FKBP-GFP-TBK1.
(J) FACS analysis of cells expressing WT FKBP-GFP-TBK1 or FKBP-GFP-TBK1 ΔC terminus (Δ 685–729), with FRB-FIs1 and mito-mKeima after 24 h of Rapalog treatment.
(K) Quantification of FACS ratiometric analysis of HeLa cells expressing FKBP-GFP-TBK1, FRB-Fis1, and mito-mKeima after 24 h of Rapalog treatment.
(L) Model of NDP52/TBK1-mediated recruitment of ULK1 complex during mitophagy resulting from bypass experiments.
All FACS quantifications: n = 3 biological replicates, except in (F) n = 2. Data are represented as mean ± SD. p value: ∗ = < 0.05; ∗∗ < 0.01; ∗∗∗ < 0.001; ∗∗∗∗ < 0.00001; ns., not significant.
See also Figures S5, S6, and S7.