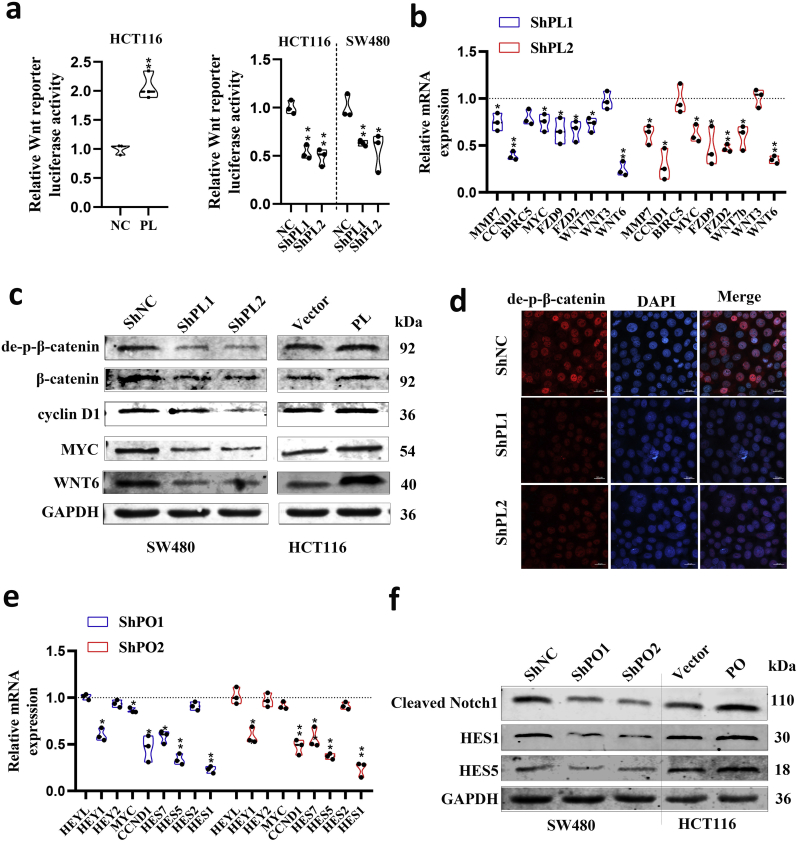
Fig. 5.
PLAGL2 and POFUT1 can activate WNT and Notch pathways, respectively.
(a)TCF reporter activity was evaluated through the β-catenin responsive TOPflash reporter in empty vector control (NC) and PLAGL2-expressing HCT116 cells or in HCT116 and SW480 cells stably transduced with lentiviral vectors carrying negative control shRNA (shNC) and PLAGL2-specific small hairpin RNAs (ShPL1 and ShPL2) *p < .05, **p < .01.
(b) qRT-PCR analyses of the expression of Wnt signaling pathway related (WNT3, WNT6, WNT7B, FZD2 and FZD9) and targeted genes (MYC, CCND1, BIRC5 and MMP7) after PLAGL2 knockdown in SW620 cells. Control group set to 1 (*p < .05, ** p < .01).
(c) Western blot analyses of related Wnt protein Wnt 6, downstream effectors of the Wnt signaling pathway including non-phospho (active) β-Catenin and total β-catenin, and accepted canonical targets including cyclin D1 and c-Myc in PLAGL2-silenced or PLAGL2-overexpressing CRC cells.
(d) Immunofluorescence of de-phospho-β -catenin in PLAGL2-silenced SW480 cells.
(e) qRT-PCR analyses of the expression of Notch signaling pathway target genes after POFUT1 knockdown in SW480 cells. Control group set to 1 (*p < .05, ** p < .01).
(f) Western blot analyses of cleaved Notch1 (the Notch1 intracellular domain), and accepted canonical targets include HES1 and HES5 in POFUT1-silenced or POFUT1-overexpressing CRC cells.