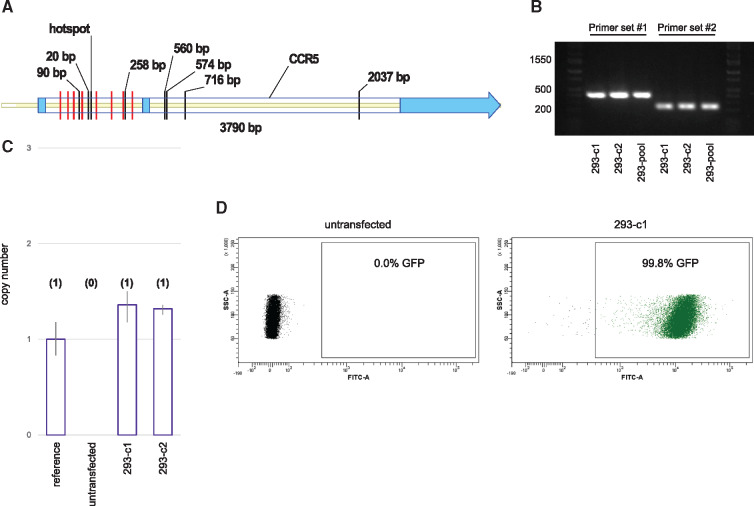
Figure 3.
RNA-guided transposition to the genome. (A) Recovered insertion sites in the CCR5 gene. Helper and donor plasmids were transfected into HEK293 cells and selected for 3 weeks. Genomic PCR was used to recover insertion sites. Ten independent insertions were found at a single TTAA hotspot. Labeled black lines indicate the location and distance from the hotspot for alternate insertion sites. Multiple independent insertions at the same TTAA for a given transfection would appear identical and be counted as a single insertion. Red lines indicate guide target sequences. CCR5 exons are shown in light blue. (B) Genomic PCR demonstrating clonal cell lines 293-c1 and 293-c2 are positive for targeted insertion to CCR5. The cell lines were derived from a positively identified well containing about 50 colonies called 293-pool. Two CCR5-directed primer sets were used. The expected sizes for a hotspot insertion using primer sets #1 and #2 were 392 and 229 bp, respectively. (C) Transposon copy number for clones 293-c1 and 293-c2. Quantitative PCR predictions were calibrated using a reference HEK293 cell line known to contain a single-copy transposon. Predicted copy number is shown in parenthesis. (D) CCR5 targeted cell lines maintained stable transgene expression following 13 weeks of culture. Flow cytometry analysis displaying GFP positive events for both untransfected HEK293 cells and an expansion of clone 293-c1.