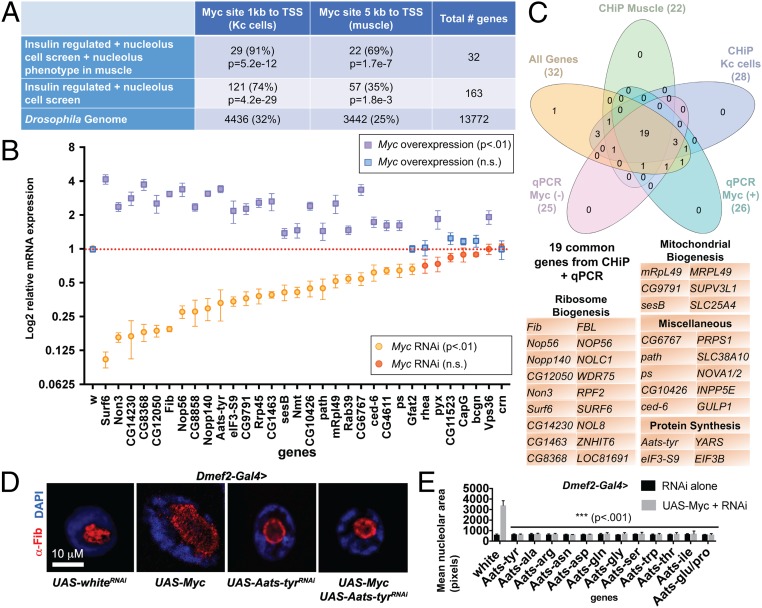
Fig. 3.
Enrichment of Myc targets reveals aaRSs as mediators of nucleolar hypertrophy. (A) Enrichment of Myc-binding sites in muscle ChIP-seq data and previously published data from Kc cells. The percentage of genes with a Myc site is lowest in the total genome, increases in the nucleolus/insulin overlap gene set, and is highest in the overlap gene set with a knockdown phenotype. P values are shown for overenrichment based on the hypergeometric distribution. (B) qPCR of putative Myc targets from Dmef2-Gal4 > UAS-MycRNAi larval muscle or Dmef2-Gal4 > UAS-Myc overexpression muscle normalized to Dmef2-Gal4 > UAS-whiteRNAi control. Significant changes in gene expression are indicated by purple squares for Myc overexpression and by orange circles for Myc knockdown. (C) Venn diagram showing overlap of the nucleolus/insulin overlap gene set with positive scoring genes from qPCR of Myc knockdown, qPCR of Myc overexpression, Myc ChIP-seq of larval muscles, and Myc ChIP-seq of Kc cells. The 19 common genes are listed. (D) Aats-tyr knockdown suppresses nucleolar hypertrophy induced by Myc overexpression. Dmef2-Gal4 was used to drive expression of UAS transgenes in larval muscle. Larvae were stained for DAPI (nucleus) in blue and α-Fibrillarin (nucleolus) in red. (E) Knockdown of 8 additional aaRS genes was able to suppress Myc-induced hypertrophy to wild-type levels. Mean area of α-Fibrillarin stain (nucleolus) from single VL4 muscles (>6 larvae) following knockdown of each gene by Dmef2-Gal4+/− UAS-Myc.