Significance
Stalling replication forks require strict cellular regulations for their stability. The intra-S phase checkpoint has been demonstrated essential for preventing stalling/stalled replication forks from collapse. In this study, a cellular pathway was uncovered that regulates the stabilization of stalling replication forks. We found that, when replication forks stall, the chromatin surrounding stalling forks becomes compacted. The fork stalling-induced chromatin compaction is achieved by modification changes on nucleosome histones, including H2BK33 deacetylation and H3K9 trimethylations. The experimental evidence further indicated that the replication fork stalling-induced chromatin compaction prevents the separation of the replicative helicase CMG and DNA polymerases, thus stabilizing stalling replication forks. This regulation pathway is named the “chromsfork” control: chromatin compaction stabilizes stalling replication forks.
Keywords: DNA replication, replication fork stability, chromatin structure, histone modification
Abstract
DNA replication forks in eukaryotic cells stall at a variety of replication barriers. Stalling forks require strict cellular regulations to prevent fork collapse. However, the mechanism underlying these cellular regulations is poorly understood. In this study, a cellular mechanism was uncovered that regulates chromatin structures to stabilize stalling forks. When replication forks stall, H2BK33, a newly identified acetylation site, is deacetylated and H3K9 trimethylated in the nucleosomes surrounding stalling forks, which results in chromatin compaction around forks. Acetylation-mimic H2BK33Q and its deacetylase clr6-1 mutations compromise this fork stalling-induced chromatin compaction, cause physical separation of replicative helicase and DNA polymerases, and significantly increase the frequency of stalling fork collapse. Furthermore, this fork stalling-induced H2BK33 deacetylation is independent of checkpoint. In summary, these results suggest that eukaryotic cells have developed a cellular mechanism that stabilizes stalling forks by targeting nucleosomes and inducing chromatin compaction around stalling forks. This mechanism is named the “Chromsfork” control: Chromatin Compaction Stabilizes Stalling Replication Forks.
As replication forks move along chromatin DNA of eukaryotic cells, they encounter a large number of replication barriers. These barriers include various secondary DNA structures, DNA lesions, chemically modified bases, tightly DNA-bound proteins, transcription machinery, and the difficult of replicating genomic regions located at rDNA genes, centromere, and telomeres (1–6). A decreased level of deoxyribonucleotide triphosphate (dNTPs) also results in fork stalling; such a situation occurs when hydroxyurea (HU) is present or in the case of aberrantly activated oncogenes (7). To prevent stalling forks from collapse and to preserve genomic integrity, stalling forks require the intra-S phase checkpoint regulations (8–11). The replisome appears to be the primary target of the checkpoint (9, 12).
In contrast to prokaryotes, DNA replication in eukaryotes takes place in the context of chromatin. The nucleosome is the fundamental subunit of chromatin. There is approximately 1 nucleosome for every 200 bp of chromatin DNA; between two adjacent nucleosome cores, there exists 20 to 60 bp of linker DNA (13). An average length of Okazaki fragments is ∼125 to 150 nucleotides (14, 15); thus, a replication fork covers a chromatin DNA region of ∼200 bp. Consequently, physical contacts should exist between the replisome and the nucleosomes that are just ahead of and behind the replisome. As a matter of fact, replisomes play a direct role in assembling nucleosomes behind the forks (16, 17). Similarly, the disassembly of nucleosomes ahead of forks must be directly caused by a moving replication fork, although the underlying molecular mechanism remains unclear. Thus, the replisome and the nucleosomes closest to the replisome have both physical and biochemical interactions. Furthermore, DNA synthesis, replication fork movement, and both disassembly and reassembly of nucleosomes ahead of and behind the replisome are precisely coordinated (18). Therefore, the nucleosomes that are closest to the replisome form a part of replication forks (Fig. 1A). This makes it highly possible that cells may also target nucleosomes and regulate chromatin structures to stabilize stalling replication forks.
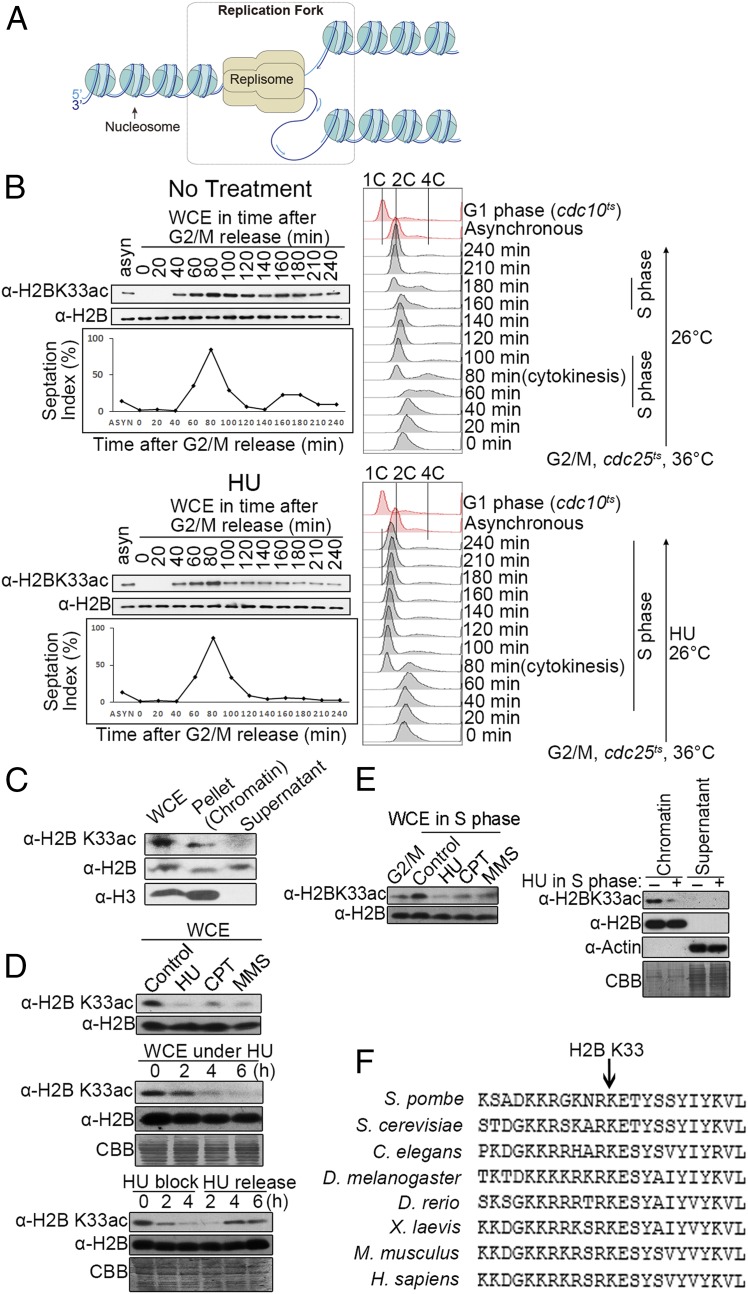
Fig. 1.
Fork stalling elicits H2BK33 deacetylation. (A) Schematic of eukaryotic DNA replication forks that are composed of replisome, nearby nucleosomes, and fork-structured DNA. (B) H2BK33 is hyperacetylated in the S phase of the cell cycle and deacetylated after fork stalling. (Top) cdc25-22 cells were released from G2/M arrest. The levels of H2B and H2BK33 acetylation were measured by Western blotting. The progression of the cell cycle was monitored by counting septa and FACS analysis. Septa appeared when the cells were in the S phase. (Bottom) cdc25-22 cells were released from G2/M arrest in the presence of 12.5 mM HU. (C) H2BK33 is acetylated in the chromatin fraction. Cell extracts were fractionated as the chromatin and supernatant (soluble) portions. The amounts of H2BK33ac, H2B, and H3 were measured by Western blotting. WCE, whole cell extracts. (D) Fork stalling causes the deacetylation of the H2BK33 site. (Top) Western blotting of WCE from asynchronized cells treated or untreated for 4 h with 12.5 mM HU, 30 μM CPT, and 0.03% MMS with a specific antibody against H2BK33ac or H2B. The levels of H2BK33ac in WCE during 12.5 mM HU block (Middle) or after HU release (Bottom) were measured by immunoblotting over time. (E) Deacetylation of the H2BK33 site in S phase cells in the presence of HU, CPT, or MMS. The cdc25-22 cells were first synchronized at G2/M and then released into the S phase in the presence of 12.5 mM HU (3 h), 30 μM CPT (3 h), or 0.03% MMS (3 h). The levels of H2BK33ac and H2B in WCE (Left) or the chromatin fraction (Right) were measured by Western blotting. (F) Alignments of a partial H2B protein sequence from the indicated species. The arrow shows the position of the S. pombe H2BK33 site.
This study found that when replication forks stall in the presence of HU, the newly identified acetylation site H2BK33 is deacetylated and H3K9 trimethylated in the nucleosomes surrounding stalling forks. These histone modifications result in a higher level of compaction in the chromatin region where replication forks stall. Next, the cellular mechanism of how fork stalling elicits chromatin compaction was examined. Furthermore, by investigating the cellular process of fork stalling-induced deacetylation of the H2BK33 site, it was investigated whether chromatin compaction is required for the stability of stalling replication forks. It was found that acetylation-mimic H2BK33Q mutation compromises fork stalling-induced chromatin compaction and results in significant instability of stalling forks. Clr6 deacetylase was identified as the enzyme responsible for deacetylating H2BK33 after fork stalling. Furthermore, it was determined that Clr6 is recruited to stalling forks by the Rad9-Hus1-Rad1 complex (9-1-1 complex). The fork stalling-induced deacetylation of H2BK33 is independent of checkpoint regulations. Impairing fork stalling-induced chromatin compaction causes the physical separation of the CMG replicative helicase and DNA polymerases in stalling forks, which results in fork collapse. Thus, eukaryotic cells have a mechanism that functions in parallel with the checkpoint and stabilizes stalling forks by targeting nucleosomes and regulating chromatin structures. This mechanism is named the “Chromsfork” control: Chromatin Compaction Stabilizes Stalling Replication Forks.
Results
Replication Fork Stalling Causes H2BK33 Deacetylation.
To test whether nucleosomes are targeted in the cellular regulation in response to replication fork stalling, histone proteins were isolated from HU-treated or untreated cells and subjected to mass spectrometry (MS) assay. In addition to detecting previously reported histone modification sites, the acetylation site H2BK33 was identified in the fission yeast Schizosaccharomyces pombe (SI Appendix, Fig. S1 A and B). Acetylation of H2BK33 was routinely detected in the chromatin fraction in normal S phase cells but hardly in HU-treated cells. This suggests that the level of H2BK33 acetylation (H2BK33ac) significantly decreased after replication forks stalled. To quantify the change in H2BK33ac levels during fork stalling, a specific antibody against a full length of S. pombe H2B was generated (SI Appendix, Fig. S1C). A specific antibody was also generated against H2BK33ac that specifically detects H2BK33ac both in vitro (SI Appendix, Fig. S1D) and in vivo (SI Appendix, Fig. S1 E–I). First, the level of H2BK33ac was measured during cell growth cycles or when replication forks stalled in the presence of HU. The results are shown in Fig. 1B. In a normal S phase in which replication forks did not stall (Upper panel), the H2BK33 site began to be acetylated when the cells were entering the S phase at ∼40 to 60 min after G2/M release. The level of H2BK33 acetylation began to increase at the beginning of S phase (∼40 to 60 min after G2/M release), reached a peak level at 80 to 100 min (S phase), and began to decrease at 120 min (the completion of S phase). The level of H2BK33ac was at its lowest at 140 min (G2 phase) and then increased again at 160 to 180 min when the cells entered into the S phase again. In the presence of HU (Fig. 1 B, Bottom), H2BK33 also began to be acetylated at the beginning of S phase (∼40 to 60 min after G2/M release); and the level of H2BK33ac reached a peak at ∼80 min. However, different from normally progressing replication forks, H2BK33 was quickly deacetylated at 100 min and kept at its lowest level in the time period of 100 to 240 min when the cells were in S phase due to HU-induced replication fork stalling. FACS analyses were carefully performed to monitor the progression of cell cycle at the indicated time points (20-min interval) after the cells were released from the G2/M arrest with or without HU presence. As shown in Fig. 1 B, Top Right (no HU), the cells had 2C DNA content at G2/M phase (0 min) (the peaks at 0–40 min were skewed to >2C, due to a large cell size for the G2/M arrested cells vs. asynchronous cells), entered into S phase at 40 to 60 min, and completed S phase at 100 min; the cells entered into a second S phase at ∼160 min and completed the S phase at ∼190 min. In the presence of HU (Fig. 1 B, Bottom Right), the cells also entered into S phase at ∼40 to 60 min and stayed in the S phase (DNA content between 1C and 2C) up to the furthest examined time point of 240 min. In fission yeast, cytokinesis is delayed and it takes place when cells are in the next round of S phase.
H2BK33 acetylation occurred in the chromatin fraction (Fig. 1C). These results suggest that H2BK33 acetylation may play an important role for DNA replication (Fig. 3B). Cells treated with HU or methyl methanesulfonate (MMS) showed significantly decreased H2BK33ac abundance (Fig. 1 D, Top), and a lower decrease of H2BK33ac level was also detected in camptothecin (CPT)-treated cells (Fig. 1 D, Top); the level of H2BK33ac drastically decreased as the time of HU treatment increased (Fig. 1 D, Middle; the whole image in SI Appendix, Fig. S1I); however, H2BK33ac was recovered after HU release (Fig. 1 D, Bottom). Furthermore, these experiments confirmed that HU-, CPT-, and MMS-induced H2BK33 deacetylation was detected in whole cell extracts and on chromatin at the S phase (Fig. 1E). This H2BK33 site is highly conserved in other eukaryotic cells (Fig. 1F).
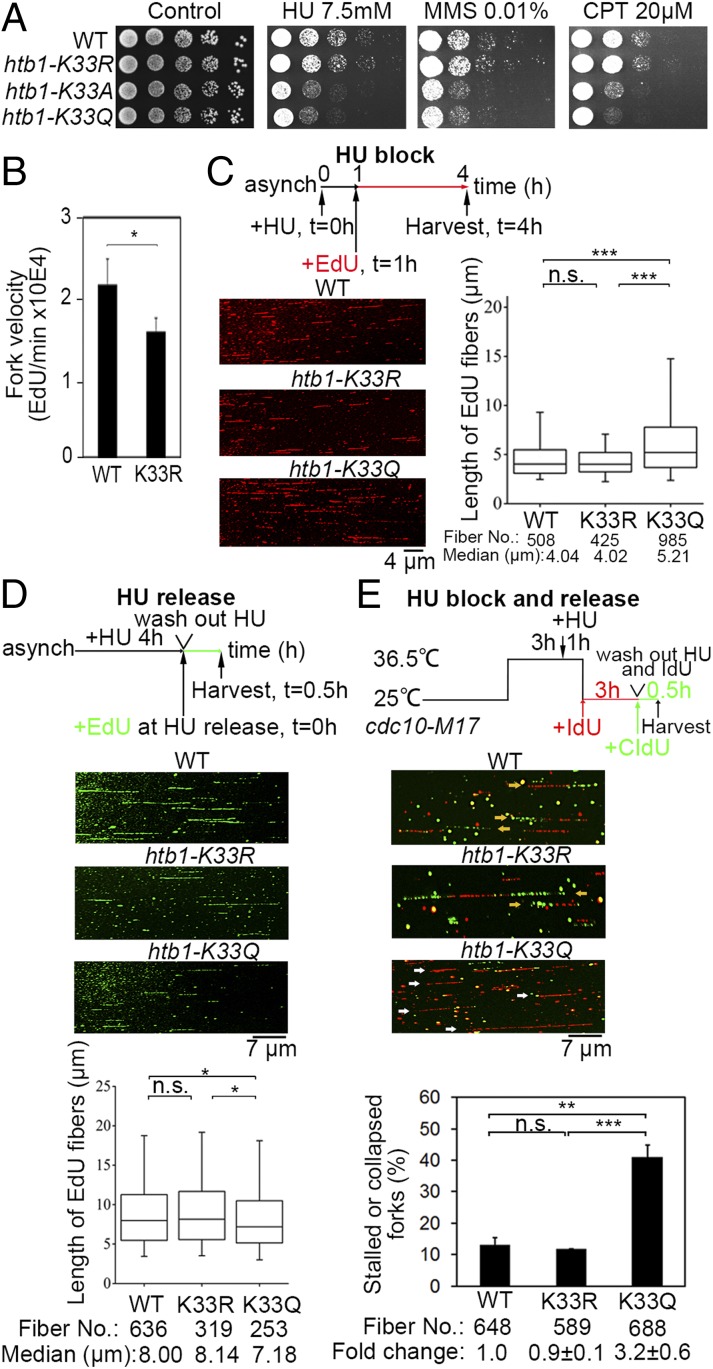
Fig. 3.
Instability of stalling replication forks in the H2BK33Q mutant. (A) Increased sensitivity of htb1-K33Q/A cells to HU, CPT, and MMS. Shown is a 5-fold serial dilution of WT and H2BK33 mutants on the indicated plates. (B) The velocity of DNA replication forks measured by FACS analysis. (C) Rate of DNA replication in the WT and H2BK33 mutants treated with HU. Asynchronized J2172 cells were first treated with 12.5 mM HU for 1 h. Then, EdU (red) was added to the culture for 3 h and the cells were harvested for the DNA combing analysis to measure the length and distribution of the EdU fibers. (D) Rate of DNA replication in the WT and H2BK33 mutants after HU release. Asynchronized cells were treated with 12.5 mM HU for 4 h. Following the removal of HU, EdU (green) was added to the culture for 0.5 h and the cells were harvested for combing analysis to measure the lengths and distributions of the EdU-incorporated DNA fibers. (C and D) Median, 25 to 75 percentiles, and 95% confidence intervals were graphed as box plots. The Mann–Whitney rank sum test was performed. (E) Percentage of stalled or collapsed forks in the WT and H2BK33 mutants during HU block and release. DNA combing assay to measure collapsed forks was performed as described in SI Appendix, Extended Experimental Procedures. Yellow arrows indicate normal forks; white arrows indicate stalled or collapsed forks. The fold change of percentages of stalled or collapsed forks in WT cells was set to 1 and expressed as mean ± SD. Statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001; P > 0.05, not significant [n.s.]) was calculated by Student’s two-tailed t test and indicated by asterisks.
H2BK33 Deacetylation and H3K9 Trimethylation around Stalling Forks.
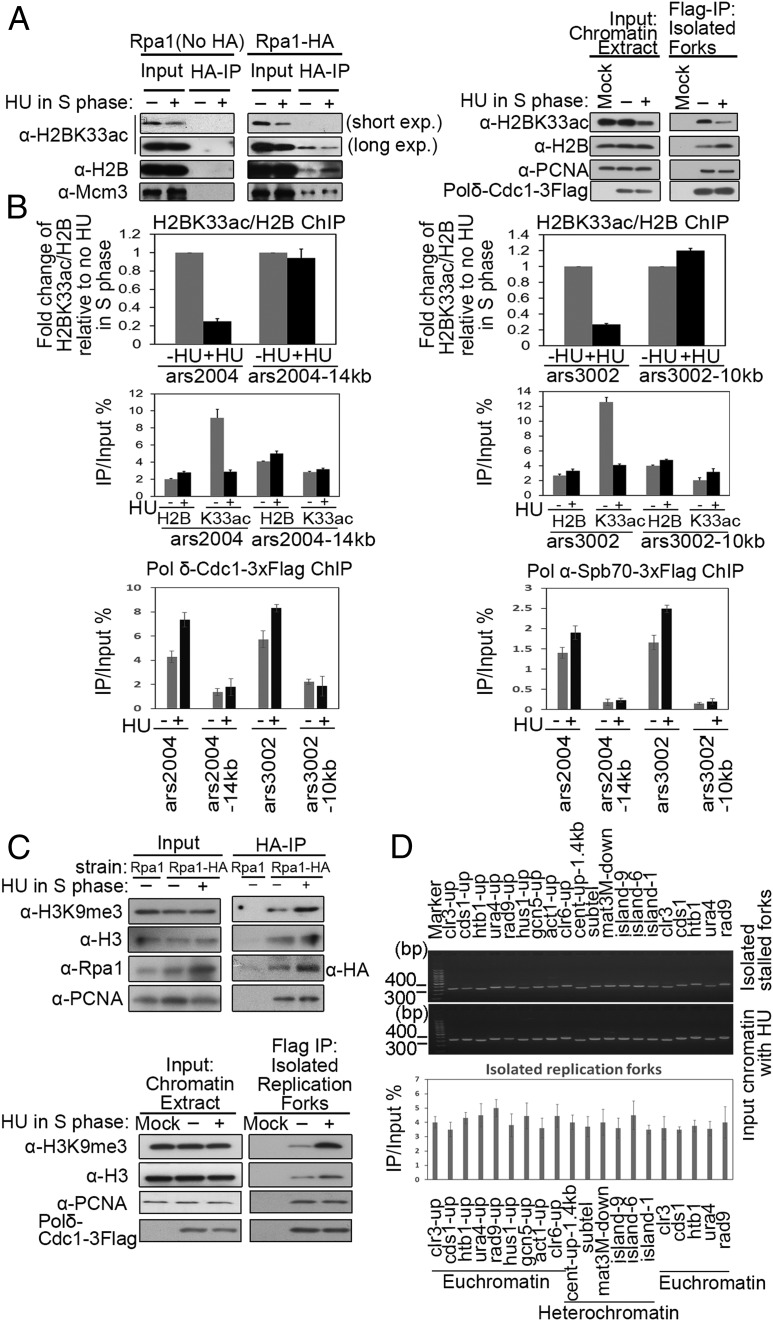
Next, the nucleosomes surrounding stalling replication forks were examined. The oligonucleosomes around the HU-stalled forks were isolated, using the HA antibody against 3HA-tagged or untagged Rpa1 (the largest subunit of RPA). RPA is a single-strand DNA binding protein that functions at DNA replication forks. As shown in Fig. 2 A, Left, it could be further confirmed that H2BK33 is deacetylated and that H2B levels are elevated at the stalling forks, although the level of H2B hardly changed in the whole chromatin (input) fraction. As a control, the amounts of Mcm3 remained unchanged (Fig. 2 A, Left). A similar result was obtained when replication forks were isolated by the immunoprecipitation against the DNA polδ-Cdc1 subunit (Fig. 2 A, Right). Furthermore, a ChIP assay also indicated that H2BK33ac was decreased after HU treatment in the S phase in the origin regions of both ars2004 and ars3002 but not in the replication fork-absent or -unstalled regions (Fig. 2B). The two Middle panels of Fig. 2B show the efficiency (as a percentage) of the DNA origin regions or the neighboring DNA regions (14 or 10 kb away from the origin) isolated via immunoprecipitation with antibody against H2BK33ac and H2B. The ChIP assay with specific antibody against 3FLAG-tagged polδ-Cdc1 subunit or polα-Spb70 subunit indicates that replication forks/replisomes were presented at the DNA origin regions of ars2004 and ars2003 but not in the neighboring DNA regions (14 or 10 kb away from the origin) (Fig. 2 B, Bottom). The specificity of ChIP with the α-H2BK33ac antibody is shown in SI Appendix, Fig. S1J.
Fig. 2.
H2BK33 deacetylation and H3K9 trimethylation occur locally at stalling replication forks. (A) Deacetylation of H2BK33 at stalled replication forks. Replication forks (replisome-fork DNA complex) were affinity purified with the indicated antibodies and the abundance of fork-associated proteins was measured by Western blotting. (B) Decreased level of H2BK33ac at stalled replication forks. The level of H2BK33ac at stalled forks was measured by ChIP-qPCR. Replisome–DNA complex was isolated as described in SI Appendix, Extended Experimental Procedures. (Top) The relative change in H2BK33ac level, which is normalized to the level of H2B, at stalled forks (+HU) versus unstalled forks (−HU). (Middle) The percentage of the indicated sites isolated in ChIP with antibody against H2B or K33ac. (Bottom) The efficiency of isolating replication forks/replisomes at the indicated sites with ChIP assays. The error bars are indicative of SD from biological replicates. (C) Trimethylation of H3K9 at stalled replication forks. The experiments were conducted as in A. The amounts of H3K9me3 and indicated factors were quantified by Western blotting. (D) Replication forks stalled randomly on chromatin. A total of 12.5 mM of HU was added to asynchronous cell cultures for 3 h. Stalling replication forks were isolated as in A or C. (Top and Middle) The amounts of DNA in the isolated replication forks or input DNA (before IP) was quantified at the indicated DNA regions by PCR and agarose gel electrophoresis. (Bottom) qPCR was used for the quantification of isolated DNA. The sequences of primers used in this study are listed in SI Appendix, Table S3.
The level of H3K9 trimethylation was also examined in the chromatin region at the stalling forks. A local chromatin region of ∼1,000 to 2,000 bp containing a stalling replication fork (HU presence) was isolated by ChIP against RPA (Rpa1) or DNA polδ-Cdc1 subunit, as shown in Fig. 2C. As a control, a similar sized chromatin region was also isolated that surrounds a normally proceeding replication fork (HU absence). The result in Fig. 2C shows that the level of H3K9me3 significantly increased in the chromatin regions surrounding the stalling replication forks. After adjusting to the amount of H3, the level of H3K9me3 at the stalling forks increased by at least 2-fold compared with unstalling forks. To confirm that the replication fork stalling induced by HU does not preferentially reside at H3K9me3-rich chromatin sites, the amounts of DNA in the isolated stalling replication forks were quantified by PCR at the chromatin sites with various levels of H3K9me3. As shown in Fig. 2D, almost equal amounts of PCR product were obtained at the 20 examined chromatin sites, 14 gene sites (euchromatin), three sites close to centromere, subtelomere, and mat3M (heterochromatin), respectively, and three island regions (facultative heterocchromatin) (19). Fig. 2D, Middle shows an equal amount of PCR products with identical sets of primers and input chromatin DNA (before ChIP). The amounts of DNA in the isolated stalling replication forks were also quantified by qPCR and showed a similar efficiency in bringing down DNA fragments at these chromatin sites (Fig. 2 D, Bottom). With the htb1-K33Q strain, an equal amount of PCR product was also obtained at euchromatin, the island, and heterochromatin regions (SI Appendix, Fig. S1K). These results indicate that the HU-induced replication fork does not preferentially stall at H3K9me3-rich chromatin sites. This is consistent with the fact that HU-induced fork stalling is caused by a lack of dNTPs rather than by chromatin structures.
Hyperacetylation of Histone H2BK33 Leads to Abnormal Fork Progression under HU and Collapse of Stalling Forks.
The mechanism of the fork stalling-induced H2BK33 deacetylation was investigated to further understand the cellular regulation of how fork stalling alters the chromatin structure and whether an altered chromatin structure then stabilizes stalling forks. First, it was examined whether the sharp deacetylation of the H2BK33 site upon replication fork stalling is critical for stalling fork stability. In S. pombe, only one copy of the htb1 gene exists for histone H2B, which is convenient for the construction of htb1 mutants. With genetic approaches, three S. pombe strains were constructed with H2BK33 mutations in the genome: htb1-K33R (no acetylation), htb1-K33Q (acetylation-mimic), and htb1-K33A (no positive charge). First, it was verified that the insertion of the selective marker kan downstream of the htb1 gene does not affect cellular growth and neither HU nor DNA damage sensitivity (SI Appendix, Fig. S2A). However, the cells that contained the acetylation-mimic H2BK33Q or H2BK33A mutations were 5- to 25-fold more sensitive to HU and MMS or CPT, compared with WT cells; however, the htb1-K33R and WT cells had comparable sensitivity to the three agents (Fig. 3A). The comparable replication stress sensitivity of H2BK33A and H2BK33Q cells suggests that the dysfunction of H2BK33 hyperacetylation is caused by the neutralization of the positive charge of K33. These results indicate that deacetylation at the H2BK33 site is critical for the stability of stalling replication forks. Furthermore, the H2BK33 mutants were not sensitive to ionizing radiation (IR), UV light (UV), mycophenolic acid (MPA, a transcription inhibitor at the elongation step), or environmental stress (high temperature [37 °C], exposure to H2O2, or 1.5 M of KCl), suggesting that H2BK33 acetylation or deacetylation is not directly involved in recombination, DNA lesion repair, or transcription (SI Appendix, Fig. S2B). In addition, H2BK33 was compared with other H2B acetylation sites in response to replication stress. H2BK5, K10, and K15 are acetylation sites in S. pombe (20). However, neither Q nor R mutation on H2BK5, K10, or K15 significantly altered the sensitivity of cells to replication stress (SI Appendix, Fig. S2C). Only when these three Q mutations were combined, a 5-fold increase in sensitivity to HU could be achieved (SI Appendix, Fig. S2C). However, synthetic sensitivity was not observed if H2BK33Q and H2BK5K10K15-3Q were combined (SI Appendix, Fig. S2D). These results indicate H2BK33 as a primary site in H2B whose acetylation is regulated in response to replication fork stalling.
While H2BK33R cells were not sensitive to HU, the velocity of replication forks was slower in H2BK33R cells compared with WT cells (SI Appendix, Fig. S2E and Fig. 3B), suggesting that H2BK33 acetylation induces a relaxed state in chromatin that makes it easier for replication forks to go through nucleosomes. Next, a single molecule-based DNA combing assay was performed to examine the stability of stalling forks in WT and htb1-K33 mutants. First, it was validated that the HU sensitivity of the WT and htb1-K33 mutants was not changed by the genetic background introduced for the DNA combing assay (SI Appendix, Fig. S2F). Then, the rate of replication fork progression was measured under the conditions of HU treatment and release according to a previously published method (21). Under HU, the median length of the EdU-labeled DNA fibers in the htb1-K33Q strain was almost 30% longer than that of the WT and htb1-K33R strains (Fig. 3C), suggesting that replication forks are more difficult to arrest in htb1-K33Q cells. In contrast, when cells were released into fresh medium after HU treatment, the median length of DNA that results from the recovery of DNA synthesis in the K33Q mutant was 11% shorter than that in WT and K33R strains (Fig. 3D). This suggests that stalling replication forks in K33Q cells either require longer time to recover and resume DNA synthesis or have more fork collapses (Fig. 3E). Furthermore, double IdU and CIdU labeling experiments (outlined in Fig. 3 E, Top) showed that the percentage of single IdU fibers, which represent unstable forks (including stalled or collapsed forks), was increased by ∼3.2-fold in K33Q cells compared with both WT and K33R cells (Fig. 3E). This result indicates that the stalling forks were not stable and collapsed at a greater rate in H2BK33Q cells. An alternative explanation for the increase of stalling fork collapse is that there is a defect to restart or repair collapsed forks in H2BK33Q cells. Although this possibility cannot be absolutely excluded, it is most likely that the fork stalling-elicited deacetylation of H2BK33 site is required for stabilizing stalling forks, because DNA recombination is involved in restarting collapsed forks but WT and H2BK33Q cells had an equal level of sensitivity to IR and UV as shown in SI Appendix, Fig. S2B. In this assay, several single CIdU tracts were also detected that represent late origin firing. The percentage of these tracts in totally labeled fibers was low and remained basically unchanged between WT and K33 mutants (SI Appendix, Fig. S2G), suggesting that the fork stalling-activated checkpoint is at a similar level (see Fig. 5). It inhibits late origin firing in both WT and K33 mutants.
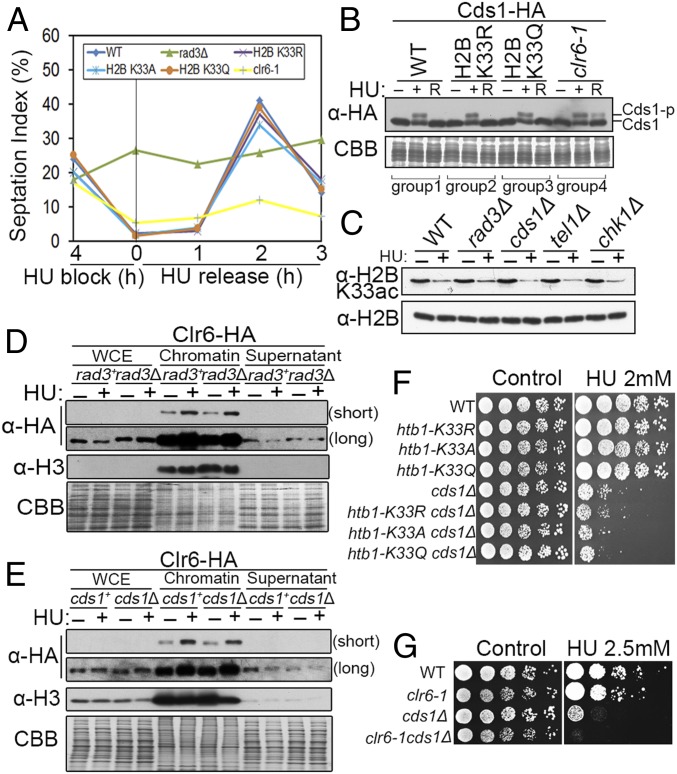
Fig. 5.
Checkpoint is intact in the clr6-1 and H2BK33 mutants. (A) Arrest of cell division by 12.5 mM HU in clr6-1 and H2BK33 mutants but not in rad3Δ cells. Cell cycle progression under HU or after HU release was monitored by the septation index. (B) Similar level of Cds1Chk2 activation by HU in the WT, H2BK33 mutants, and clr6-1 cells. HU release resulted in inactivation of Cds1 in the WT and H2BK33 mutants but not in clr6-1 cells. The phosphorylated Cds1 (Cds1-P) migrated more slowly than unphosphorylated Cds1 in SDS/PAGE gel with Phos-tag. (C) Similar level of H2BK33 deacetylation in the WT, rad3Δ, cds1Δ, tel1Δ, and chk1Δ cells under HU. (D and E) Recruitment of Clr6 onto chromatin under HU was normal in rad3Δ (D) and cds1Δ (E). The Western blotting results are shown. (F) Comparison of HU sensitivity of cds1Δ, H2BK33 mutants, and double-mutant cells. (G) Comparison of HU sensitivity of clr6-1, cds1Δ, or double-mutant cells. A 5-fold serial dilution test was performed for F and G.
Histone Deacetylase Clr6 Deacetylates H2BK33 at Stalling Replication Forks.
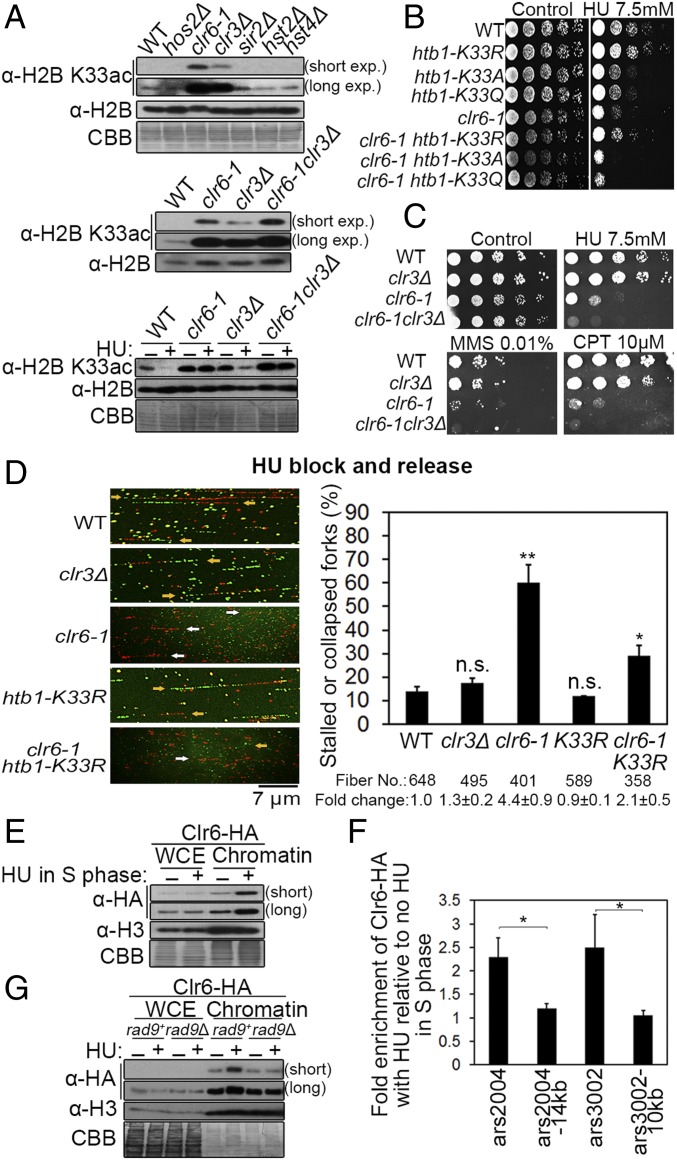
To identify the histone deacetylases (HDACs) responsible for reducing H2BK33 acetylation, the level of H2BK33 acetylation was screened in these HDAC mutants. In clr6-1 (a temperature-sensitive mutant) and clr3Δ mutants, the abundance of H2BK33ac increased and higher levels of H2BK33ac were found in clr6-1 than in clr3Δ (Fig. 4 A, Top). The clr6-1 and clr3Δ double mutant exhibited a more pronounced increase in H2BK33 acetylation than either single mutant (Fig. 4A, Middle). Furthermore, it was investigated whether Clr6 or Clr3 is required for the HU-induced deacetylation of H2BK33. The deacetylation of H2BK33 by HU treatment of clr6-1 and clr6-1clr3Δ mutants was nearly absent compared with that in the WT or clr3Δ cells (Fig. 4 A, Bottom). These results indicate Clr6 as the major deacetylase for the H2BK33 site under both physiological and replication stress conditions. A previous study also suggested Clr6 to have a higher level of activity and a broad range of substrates compared with Clr3 (22).
Fig. 4.
Clr6 is identified as the primary deacetylase of H2BK33. (A, Top) Level of H2BK33 acetylation in the 6 HDACs (histone deacetylases)-defective S. pombe mutants. clr6-1 is a temperature-sensitive mutant. The effects of clr3Δ, clr6-1, and clr3Δ-clr6-1 double mutations on H2BK33 acetylation are shown under unperturbed growth conditions (A, Middle) or HU treatment (A, Bottom). (B) Comparison of growth sensitivity to HU of the WT, clr6-1, htb1-K33R/A/Q mutants, and the double mutants. (C) Growth sensitivity of the clr6 and clr3 mutants to HU, CPT, and MMS. (D) DNA combing analysis of fork stalling or collapse in WT, clr3Δ, clr6-1, htb1-K33R, or clr6-1K33R cells. Representative DNA fibers are shown (yellow arrows indicate normal forks; white arrows indicate stalled or collapsed forks). The total number of calculated DNA fibers and the fold change of percentages of stalled or collapsed forks are shown below the plot. (E) Recruitment of Clr6 onto chromatin is enhanced under HU treatment. (F) ChIP-qPCR analysis of Clr6 loading onto the stalled replication forks at ars2004, ars3002, and neighboring regions under HU. The level of Clr6 without HU treatment is assumed to be “1”. (G) Requirement of Rad9 for the recruitment of Clr6 onto chromatin under replication fork stalling. *P < 0.05; **P < 0.01; ***P < 0.001; P > 0.05, not significant (n.s.).
The clr6-1 cells were highly sensitive to HU treatment even at 32 °C (semipermissive temperature) (Fig. 4 B and C), which is consistent with a previous result (23). To further confirm that H2BK33 is a substrate of Clr6 in vivo, clr6-1 and htb1-K33 double mutants were constructed and their HU sensitivities were compared. As shown in Fig. 4B, clr6-1 htb1-K33R cells were less sensitive to HU than clr6-1. This result supports the biochemical data indicating that Clr6 deacetylases H2BK33 (Fig. 4A). Why did the H2BK33R mutation not completely rescue the sensitivity of clr6-1 cells to HU? The answer is probably that Clr6 could also deacetylate some other histone sites for chromatin compaction and stabilization of stalling replication forks. The clr6-1 htb1-K33Q/A cells were slightly more sensitive to HU than clr6-1. The reason is that Clr6-1 maintains some residual activity at a semipermissive temperature that deacetylates the H2BK33 site for stabilizing stalling replication forks to a certain extent. Furthermore, the clr6-1 mutant displayed a more severe growth defect than clr3Δ under HU, CPT, and MMS treatments. A slightly synthetic defective phenotype was also observed in the clr6-1clr3Δ double mutant (Fig. 4C), which is consistent with Clr6 being the major deacetylase for the H2BK33 site.
A DNA combing assay was used to examine the stability of stalling forks in WT, K33R, clr3Δ, clr6-1, and clr6-1K33R mutants. The genetic background introduced for the DNA combing assay did not change the HU sensitivity of WT, clr3Δ, and clr6-1 mutants (SI Appendix, Fig. S3A). Consistent with its HU sensitivity, a DNA combing assay, with a similar procedure as outlined in Fig. 3E, showed that the clr6-1 mutant exhibits ∼4.4- and 3.4-fold increases in stalled or collapsed forks compared with WT and the clr3Δ mutant, respectively (Fig. 4D). The introduction of the K33R mutation into the clr6-1 decreased the rate of stalling fork collapse (Fig. 4D), which is consistent with the finding that clr6-1 htb1-K33R cells were less sensitive to HU than clr6-1 (Fig. 4B). Again, the rate of stalling fork collapse was slightly lower in the K33R cells compared with WT (Fig. 4D). The results in Fig. 4 E and F and SI Appendix, Fig. S3C show that Clr6 was enriched on chromatin and locally enriched in the stalling forks in the presence of HU, although the total cellular amount of Clr6 remained unchanged. Unlike Clr6, Clr3 was only slightly enriched in stalled forks (SI Appendix, Fig. S3B). To determine how Clr6 is recruited onto chromatin after fork stalling, Clr6 interacting proteins were searched and the 9-1-1 complex (Rad9-Rad1-Hus1) was found via immunoprecipitation. The interaction between Clr6 and Rad9 was confirmed by co-IP in the presence or absence of DNase I (SI Appendix, Fig. S3D). Also, the yeast two-hybrid assay confirmed the interaction between Clr6 and the 9-1-1 complex (SI Appendix, Fig. S3E), and a strong interaction was detected between Clr6 and Hus1 or the Rad9 subunit of the 9-1-1 complex (SI Appendix, Fig. S3E). This is consistent with previous reports that indicated the interaction of HDA-3 or HDAC1 (Clr6 homolog) and the 9-1-1 complex in Caenorhabditis elegans and humans, respectively (24). This study further demonstrated that Rad9 (a subunit of the 9-1-1 complex [Rad9-Hus1-Rad1]) is required for the recruitment of Clr6 onto chromatin in response to the presence of HU (Fig. 4G).
Clr6-Mediated H2BK33 Deacetylation Is Independent of Checkpoint Regulation.
Next, it was examined whether H2BK33 deacetylation is related to checkpoint regulation. First, it was examined whether H2BK33 mutants affect checkpoint activation. The cell cycle progression arrests when the checkpoint is activated by replication fork stalling; thus, this assay can be used to check the integrity of the checkpoint. The cell cycle progression in H2BK33 mutants, clr6-1, and the WT were blocked in the presence of HU but not in rad3Δ cells (Rad3 is equivalent to ATR in S. pombe) (Fig. 5A). After HU release, cell division continued in the H2BK33 mutants as evidenced by changes in the septation index. However, only a small percentage of clr6-1 cells returned to cell cycle (Fig. 5A). In support of this result, HU-induced Cds1Chk2 activation (Cds1 phosphorylation) was normal in H2BK33 and clr6-1 mutants compared with WT (Fig. 5B). After HU release, Cds1 was inactivated as shown by its dephosphorylation (the Cds1 band moved faster than the Cds1-p) in the WT and H2BK33 mutants. However, a significant portion of Cds1 remained activated in clr6-1 cells after HU release (Fig. 5B), which implies that severe DNA damage occurred in clr6-1 cells during fork stalling. This result explains why the clr6-1 cells did not return to the cell cycle after HU release (Fig. 5A). This result is also consistent with the ∼40% higher rate of stalling fork collapse in the HU-treated clr6-1 cells than that in the H2BK33Q mutant (Figs. 3E and 4D). Furthermore, HU-induced H2BK33 deacetylation remained unaffected in the absence of Rad3ATR, Cds1Chk2, Tel1ATM, or Chk1 kinase (Fig. 5C). This result was supported by the unaffected recruitment of Clr6 to chromatin following HU treatment in the absence of either Rad3 or Cds1 (Fig. 5 D and E). Additionally, epistasis analysis between cds1Δ and H2BK33 mutants showed that the H2BK33R mutation did not rescue the sensitivity of cds1Δ cells to HU, while synthetic HU sensitivity was detectable in cds1Δ htb1-K33Q/A cells (Fig. 5F). Synthetic HU sensitivity was also observed in the clr6-1 cds1Δ cells (Fig. 5G). Taken together, these results indicate that (i) checkpoint activation is unaffected in H2BK33 and clr6-1 mutants and (ii) the instability of stalling forks in H2BK33Q/A and clr6-1 cells under HU treatment is not caused by checkpoint dysfunction. Thus, fork stalling-induced H2BK33 deacetylation and intra-S phase checkpoint are two parallel pathways.
H2BK33Q and clr6-1 Mutations Result in Relaxed Chromatin and Genomic Instability at the rDNA Regions.
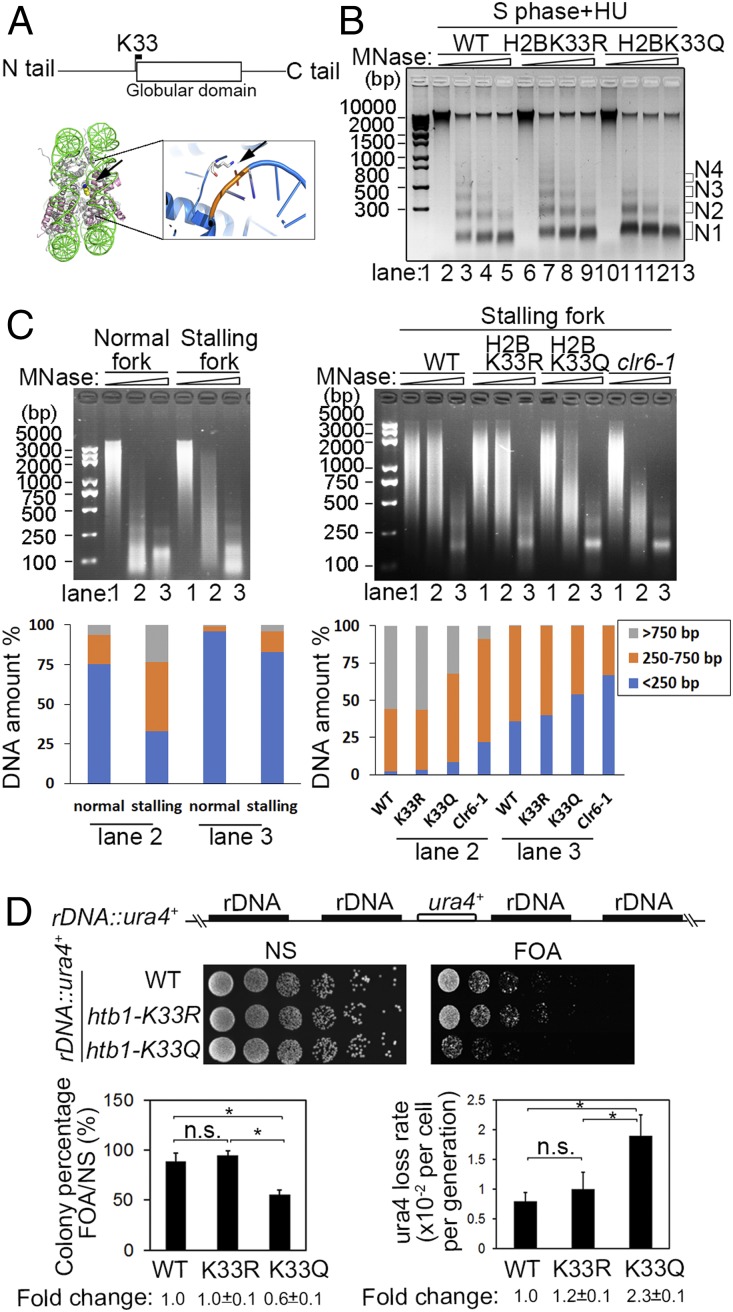
Histone acetylation is generally associated with gene activation and chromatin relaxation (25). Replication fork stalling simultaneously induced H2BK33 deacetylation and H3K9 trimethylation in the nucleosome around the stalling forks (Fig. 2 A–C). This suggests that H2BK33 deacetylation may be associated with chromatin compaction. Based on the crystal structure of nucleosomes (26), the H2BK33 residue is close to the globular domain (Fig. 6A). The positive charge of the H2BK33 ε-amino group on the lateral surface of the nucleosome is in direct contact with the negative charge of the phosphate group of the intranucleosome DNA phosphodiester backbone (Fig. 6A). The K33 residue resides within a stretch of 8 basic residues and the H2B repression (HBR) motif that is implicated in the facilitation of internucleosome interactions (26–28). Therefore, H2BK33 deacetylation should enhance nucleosome–DNA interaction and induce a compacted chromatin structure, thus impeding replication forks to go through such a chromatin structure. A slower fork speed was detected in the H2BK33R mutation where the acetylation of this site is blocked (Fig. 3B). To obtain direct evidence that H2BK33 deacetylation, induced by replication fork stalling, increases chromatin compaction, the MNase digestion assay was performed. The chromatin in K33Q cells was digested more readily than in the WT and K33R mutant in the S phase and under HU conditions (Fig. 6B). These results indicate that H2BK33 acetylation relaxes chromatin for replication forks to go through nucleosomes more easily; however, the deacetylation of H2BK33 plays a critical role in the formation of chromatin compaction elicited by replication fork stalling. The MNase digestion assay was also performed on the S phase of chromatin in the cells treated or untreated by HU. The result in SI Appendix, Fig. S4A indicates that chromatin became more resistant to MNase digestion as the time of HU treatment increased. The experiment in SI Appendix, Fig. S4A was an assay of whole chromatin resistance to MNase. The detection of MNase resistance of whole chromatin suggests that a significant portion of the chromatin region around each stalling fork is compacted. A MNase digestion assay on the local chromatin region of stalling replication forks showed much stronger resistance to MNase digestion than on the chromatin regions surrounding normal replication forks (Fig. 6 C, Left). As expected, the chromatin regions surrounding stalling replication forks in K33Q and clr6-1 cells was digested more readily than in the WT and K33R mutants (Fig. 6 C, Right). The amount of DNA at the indicated range of DNA size was quantified and shown in Fig. 6 C, Lower. Taken together, these results indicate that the chromatin regions around stalling forks are highly compacted.
Fig. 6.
A relaxed chromatin and genomic instability at the native rDNA replication barrier site in the H2BK33Q mutant cells. (A and B) Chromatin compaction is compromised in H2BK33Q mutant cells. (A, Top) Location of K33 in the histone H2B sequence. (A, Bottom) H2BK33 indicated by arrow contacts with DNA within the nucleosome core particle (Xenopus laevis, PDB 1AOI), and this interaction is enlarged on the Right. (B) MNase digestion of chromatin from the WT, H2BK33R, and H2BK33Q cells under HU treatment. The cdc25-22 cells were first arrested at G2/M phase and then released into S phase in the presence of HU. N1-4 indicates the number of nucleosomes. (C) MNase digestion of local chromatin regions of normal or stalling replication forks. The cdc25-22 Polδ-cdc1-3Flag cells with various genetic background indicated below were first arrested at G2/M phase and then released into S phase in the absence or presence of HU. The experimental method is basically similar to Fig. 2 A, Right except that chromatin was sonicated to an average size of 2,000 to 5,000 bp. (Left) The WT cells arrested at G2/M phase were released into S phase in the absence or presence of HU. (Right) WT, H2BK33R, H2BK33Q, and clr6-1 cells were released into S phase in the presence of HU. (Bottom) The percentage of DNA amount for the indicated range of DNA size was measured for lanes 2 and 3 by Quantity One software. (D) A relaxed chromatin results in genomic instability in the rDNA genomic locus in htb1-K33Q cells. (Top) ura4 gene integrated into the spacer region in the rDNA genes in the WT, htb1-K33R, and htb1-K33Q strains. (Middle) Examination of expression level of ura4 gene at the rDNA locus in the WT and H2BK33 mutants by spot assay. (Bottom Left) The percentage of colonies grown in FOA plates. (Bottom Right) Loss rate of the ura4 gene in WT and H2BK33R/Q mutant cells. *P < 0.05; P > 0.05, not significant (n.s.).
The H2BK33Q mutation resulted in an increased rate of stalling replication fork collapse (Fig. 3E), consequently causing genomic instability. To directly test whether genomic regions at native replication barriers become more unstable in H2BK33Q mutant cells, three strains were constructed with a copy of the ura4 gene inserted into the spacer region between the rDNA genes in WT and H2BK33R/Q cells (the native ura4 gene is defective due to DS/E truncation in these cells) (Fig. 6 D, Top). Each rDNA repeat of 10.9 kb contains at least 8 natural replication fork barriers, including 5 G4 motifs and 3 Ter1-3 sites that stall replication forks either by blocking DNA polymerases or the replicative helicase (1, 4–6, 29). First, the expression level of the ura4 gene was examined in FOA (5-fluorouracil-6-carboxylic acid monohydrate; 5-FOA) plates. SI Appendix, Fig. S4B shows this assay as a control, where ura4Δ cells grew best in the FOA plate compared with ura4+ (normal expression, no growth) and rDNA::ura4+ cells (decreased expression, some growth). As expected, the expression of the ura4 gene was increased in htb1-K33Q cells compared with WT, because a 5-fold dilution assay showed that the growth of htb1-K33Q cells was noticeably inhibited on the FOA plate (Fig. 6 D, Top). The growth difference on the FOA plate was not significant between htb1-K33R and WT cells (Fig. 6 D, Top). An assay of the survival rate on FOA plates further indicated that htb1-K33Q cells had the worst growth rate among all 3 strains (Fig. 6 D, Bottom Left). These results are consistent with htb1-K33Q cells that have more relaxed chromatin compared with WT and htb1-K33R cells (Fig. 6B).
As shown in Fig. 6 D, Bottom Right, the loss rate of the ura4 gene was nearly 2-fold higher in H2BK33Q cells than in WT and H2BK33R cells. PCR analysis showed that the ura4− phenotype was primarily a result of the loss of the entire ura4 gene (SI Appendix, Fig. S4C). This result indicates that the genomic locus at the native replication barrier site becomes more unstable in H2BK33Q cells where fork stalling-elicited chromatin compaction is compromised and stalling forks collapse at a higher rate.
Impairing Fork Stalling-Induced Chromatin Compaction Causes Physical Separation of DNA Polymerases and CMG Replicative Helicase.
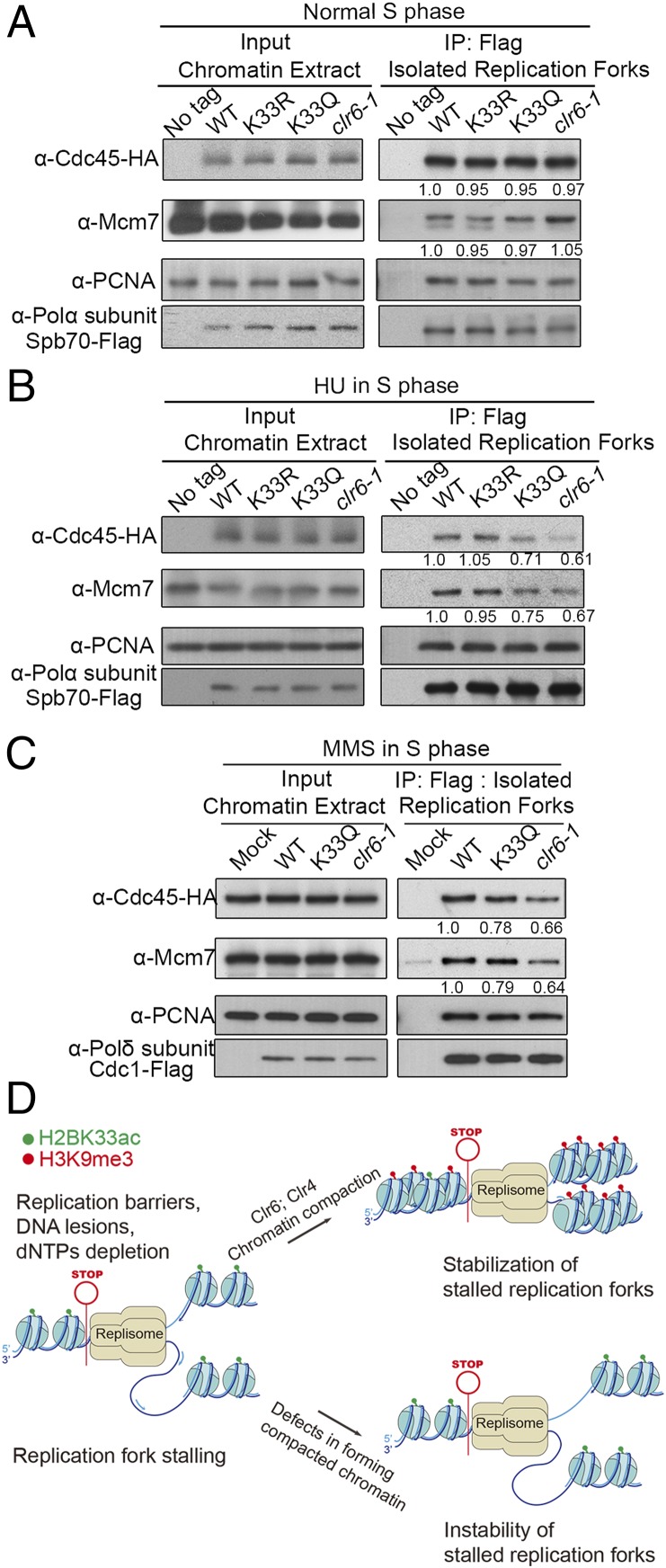
The replisome is a macromolecular complex comprised of approximately 2 to 3 dozen proteins/protein complexes, which is the central component of replication forks. Therefore, maintaining replisome integrity must be a key mechanism to stabilize the stalling replication forks. Two key biochemical reactions occur in replication forks: the first one is the DNA replicative helicase-mediated unwinding of template DNA; the second one is the coordinated synthesis of leading and lagging strands catalyzed by DNA polymerases α, δ, and ε. Both biochemical reactions must be precisely coordinated to fulfill semiconservative DNA replication on double-strand DNA. However, this precise coordination could be disrupted via replication fork stalling, which results in replication fork collapse. Thus, to explore why the chromatin compaction around stalling replication forks is required to prevent fork collapse, we examined whether the association of DNA replicative helicase CMG (Cdc45-MCM-GINS) and DNA polymerases-DNA polymerase α and δ is altered when fork stalling-induced chromatin compaction is compromised. H2BK33R/Q or clr6-1 mutations were introduced into the strain that contains HA-tagged Cdc45 and FLAG-tagged DNA polymerase α-subunit Spb70 or pol δ-subunit Cdc1 and it was verified that the HU sensitivity did not change by those tagged proteins (SI Appendix, Fig. S4D). When replication forks were not stalling, the levels of DNA polymerase α (Spb70 subunit) and CMG (Cdc45 subunit) on chromatin or in isolated replication forks were similar in WT, K33R, K33Q, and clr6-1 mutants (Fig. 7A). When replication forks were stalled in the presence of HU, the levels of Spb70, Mcm7 (a subunit of MCM), and Cdc45 on chromatin were still similar in the WT, K33R, K33Q, and clr6-1 cells (Fig. 7B), indicating that the CMG helicase and DNA pol α still associate with chromatin. However, in isolated forks, the levels of Spb70 remained comparable among cells, while the levels of Cdc45 and Mcm7 decreased remarkably by ∼30% and ∼40%, respectively, in the K33Q and clr6-1 cells. This indicates that, in a significant portion of stalling forks, the CMG helicase separated from replication forks (Fig. 7B). This separation of the CMG helicase from DNA polymerase δ or replication forks was also observed in the K33Q and clr6-1 mutants, when replication forks were stalled by the presence of MMS (Fig. 7C). These results indicate that replication fork stalling-induced chromatin compaction prevents the separation between the CMG replicative helicase and DNA polymerases in stalling replication forks. Thus, the collapse of stalling replication forks was prevented.
Fig. 7.
Replication fork stalling-induced chromatin compaction prevents physical separation of DNA polymerases and the CMG replication helicase. Chromatin compaction prevents separation of DNA polymerase α or δ and the CMG helicase. Formaldehyde cross-linked chromatin was sonicated to an average size of ∼700 bp. Replisomes were isolated by ChIP with monoclonal FLAG antibody against Spb70-FLAG or Cdc1-3Flag. (A) The levels of Cdc45-HA and Spb70-FLAG on chromatin and isolated replication forks in normal S phase. The cells with cdc25-22 background were first arrested at G2/M phase and then released to S phase 90 min after G2/M release. (B) The levels of Cdc45-HA, MCM (Mcm7 subunit), and Polα-Spb70-FLAG on chromatin and isolated replication forks in HU-treated cells of S phase. As in A, the cells were first arrested at G2/M phase and then released to S phase under HU treatment of 3.5 h. (C) The levels of Cdc45-HA, MCM (Mcm7 subunit), and Polδ-Cdc1-FLAG on chromatin and isolated replication forks in MMS-treated cells of S phase. The assay was performed as in B. (D) Diagram of the chromsfork control to stabilize stalling replication forks. When DNA replication forks encounter replication barriers and stall, chromatin compaction around stalling forks is elicited through altering histone modifications, including H2BK33 deacetylation, H3K9 trimethylation, and increasing nucleosomal density. A compacted chromatin prevents physical separation of DNA polymerases and the replicative CMG helicase. This stabilizes stalling replication forks.
Discussion
This study discovered a cellular mechanism that is required for the stability of stalling replication forks. This mechanism is named the “chromsfork” control; replication fork stalling elicits chromatin compaction, and the resulting chromatin compaction is crucial for preventing stalling fork collapse (Fig. 7D). Previous studies have demonstrated that the intra-S phase checkpoint is an essential regulation step for stabilizing stalling forks (1, 8–11). Thus, at least two major independent mechanisms exist in eukaryotic cells for the stability of stalling replication forks: the intra-S phase checkpoint and the chromsfork control.
A comparison of the chromsfork control with the S phase checkpoint shows that both are activated by replication fork stalling and effect their actions back to forks for their stability. However, fundamental differences exist between the two mechanisms. First, the S phase checkpoint mainly uses ATR and Chk2 (the functional homolog of yeast Chk2 is Chk1 in metazoans) protein kinases to both regulate and control the functions of replisome components (30). However, the histone deacetylases and methylases are suggested to be among the primary enzymes required to achieve chromatin compaction in the chromsfork control (Fig. 7D) (13, 31–33). Second, the principle targets of the S phase checkpoint appear to be replisome components and several nonreplisome proteins (9, 12, 34–37). However, in the chromsfork control, nucleosomes are primarily targeted and regulated, which leads to more compacted chromatin surrounding stalling forks through altering histone modifications (Fig. 7D). We suggest that these fundamental differences provide the basis for the parallel function of the intra-S phase checkpoint and the chromsfork control for stabilizing stalling replication forks.
The discovery of the chromsfork control provides a mechanistic explanation for why silencing factors are required for the maintenance of genomic stability in the difficult-to-replicate genomic loci. A study in Drosophila indicated that a functional defect in the genes Su(var)3–9 (H3K9 methyl transferase) or dcr (dicer-2) destabilizes rDNA and satellite DNA regions (38). A further relevant study showed that a defective Stw1 results in a reduced level of trimethylated H3K27 and H3K9, causing an elevated level of DNA break (39). Both rDNA genes and satellite sites harbor replication barriers (3–5, 29). Therefore, fork stalling at these regions requires silencing factors to elicit chromatin compaction to prevent stalling fork collapse and to maintain genomic integrity in these difficult-to-replicate genomic regions. The present finding that a compromised fork stalling-induced chromatin compaction in the H2BK33Q cells resulted in genomic instability in the rDNA region (Fig. 6C) is consistent with the above-mentioned two findings of previous studies. The chromsfork control may also explain two previous observations: tight protein–DNA interactions favor gene silencing (40) and DNA replication causes heterochromatin spreading at the silent mating-type loci in budding yeast (41). A replication barrier site exists at the yeast mating-type locus (1); thus, replication fork stalling at this site causes chromatin compaction or heterochromatin spreading. In the study of Dubarry et al. (40), an array of 256 lac operators was integrated 1.5 kb upstream of the ADE2 gene. Very likely, replication forks stall at the extensive array of lac operators, which induces chromatin compaction and subsequently results in gene silencing. Dubarry et al. (40) also reported that Sir3 and Sir4 (two subunits in the Sir2 deacetylase complex) are genome-wide enriched at natural replication pause sites, which supports the findings of this study that Clr6 deacetylase-mediated deacetylation of H2BK33 plays a critical role in the chromsfork control (Fig. 7D).
In contrast to a moving replication fork that requires the frontal chromatin structure to be relaxed, a stalling or stalled fork requires the chromatin surrounding to be compacted for its stabilization. The reason will likely be relevant for the fundamental process of DNA replication. Two central biochemical reactions exist in replication forks: One is the unwinding of dsDNA, which is catalyzed by the replicative helicase; the other is the synthesis of leading and lagging strands catalyzed by DNA polymerases α, δ, and ε. Both the unwinding of dsDNA and DNA synthesis at replication forks must be strictly coordinated. In other words, the biochemical actions of the replicative helicase and DNA polymerases must be coupled in replication forks. Replication fork stalling can either be caused by blocking the replicative helicase or by impeding DNA polymerases. DNA polymerases have an extremely fine structure in their catalytic center, which ensures the extremely high fidelity of DNA synthesis. For example, the active center in DNA polymerases can accommodate A:T or G:C pairs but not A:C and G:T mismatches (42). Although DNA helicases are stopped when they encounter a tightly DNA-bound protein, secondary DNA structures, or a bulky chemical group (e.g., a protein or a long peptide chain) covalently linked to a base, DNA polymerases can be more easily impeded via chemically modified bases, base lesions, secondary DNA structures formed in the DNA template for lagging strand synthesis, and the absence of dNTPs. When DNA polymerases are impeded, the movement of the replicative helicase must be stopped to prevent it from leaving. A compacted chromatin structure ahead of stalling forks should provide resistance to the replicative helicase moving away from stalling forks or DNA polymerases. Thus, stalling replication forks are stabilized. The cellular regulation of H2BK33 acetylation or deacetylation provides a good example in the Chromsfork control for stabilizing stalling replication forks. For normally progressing replication forks, the chromatin region ahead of the replication fork is required to be at a relaxed state. Thus, replication forks can go through nucleosomes. It was found that H2BK33 acetylation is highest at the S phase during cell division cycles (Fig. 1 B, Top). This result, together with the result shown in Fig. 3B, suggests that H2BK33 acetylation plays a critical role in relaxing chromatin for replication forks moving through nucleosomes. In the presence of HU, replication forks are stalling due to a reduced level of dNTPs. In this case, fork stalling is caused by impeding DNA polymerases. Impeding DNA polymerase will potentially disrupt the required coordination between the biochemical actions of the replicative DNA helicase and DNA polymerases. To prevent the replicative helicase from moving away from replication forks when DNA polymerases are impeded, the chromatin regions surrounding stalling forks are made more compacted through precise cellular regulations that result in H2BK33 deacetylating, H3K9 trimethylation, and maybe some other not-yet-identified histone modifications or protein factors. A compacted chromatin surrounding stalling replication forks provides resistance to the replicative helicase moving away from forks. Thus, stalling forks are stabilized. However, in the H2BK33Q cells, the chromatin compaction surrounding stalling forks is compromised, resulting in a fast moving fork (less prone to stall) in the presence of HU (Fig. 3C). A low degree of chromatin compaction has less resistance to the replicative helicase moving away from forks/DNA polymerases, and this results in more collapse of stalling forks (Fig. 6 C, Right and Fig. 7 B and C).
The replication fork stalling-induced chromatin compaction may be related to the formation of heterochromatin domains. In fission yeast, there are four heterochromatin regions: rDNA genes, mating-type loci, centromere regions, and telomere regions. All four genomic regions have distinct DNA sequences, and the proteins associated with these sequences are also very different. Previous studies have reported that noncoding RNA is involved in the formation of heterochromatin domains (43–46). Further studies indicated that centromere, mating-type locus, and telomere have a common sequence that is bound by RNAi-RITS and relevant to the formation of heterochromatin (47, 48). However, all four regions also have two things in common: the heterochromatin structure and the harboring of a DNA replication barrier(s). DNA replication barriers in these genomic regions stall replication forks and subsequently elicit chromatin compaction, which is suggested to be one of the critical factors that cause the development of the heterochromatin structure.
Materials and Methods
The information of strains, plasmids, antibody generation, preparation of cell or chromatin extracts, DNA combing assay, isolation of replication forks, MNase assay, and ChIP-qPCR are in SI Appendix.
Supplementary Material
Acknowledgments
We thank the members of the D.K. laboratory, Yuze Gao, Yang Liu, Lilin Du, Antony Carr, and Karl Ekwall for help and pombe strains. This work was supported by grants from the Ministry of Science and Technology of China (2013CB911000 and 2016YFA0500301), the National Natural Science Foundation of China (31661143032, 31230021, and 31730022), the Peking-Tsinghua Center for Life Sciences, and the National Key Laboratory of Protein and Plant Gene Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821475116/-/DCSupplemental.
References
- 1.Mizuno K., Miyabe I., Schalbetter S. A., Carr A. M., Murray J. M., Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493, 246–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres J. Z., Schnakenberg S. L., Zakian V. A., Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24, 3198–3212 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirkin E. V., Mirkin S. M., Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71, 13–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabouri N., Capra J. A., Zakian V. A., The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 12, 101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallgren M., et al. , G-rich telomeric and ribosomal DNA sequences from the fission yeast genome form stable G-quadruplex DNA structures in vitro and are unwound by the Pfh1 DNA helicase. Nucleic Acids Res. 44, 6213–6231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahan D., et al. , Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures. Nucleic Acids Res. 46, 11847–11857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bester A. C., et al. , Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sogo J. M., Lopes M., Foiani M., Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Hu J., et al. , The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149, 1221–1232 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Tercero J. A., Diffley J. F., Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412, 553–557 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Cha R. S., Kleckner N., ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297, 602–606 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Katou Y., et al. , S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Kornberg R. D., Lorch Y., Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Smith D. J., Whitehouse I., Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B., Hu J., Wang J., Kong D., Direct visualization of RNA-DNA primer removal from okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J. Biol. Chem. 292, 4777–4788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibahara K., Stillman B., Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Liu S., et al. , RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science 355, 415–420 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Prado F., Maya D., Regulation of replication fork advance and stability by nucleosome assembly. Genes (Basel) 8, E49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allshire R. C., Ekwall K., Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 7, a018770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong L., Wang Y., Mapping post-translational modifications of histones H2A, H2B and H4 in Schizosaccharomyces pombe. Int. J. Mass Spectrom. 301, 159–165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatinos S. A., Green M. D., Forsburg S. L., Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol. Cell. Biol. 32, 4986–4997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjerling P., et al. , Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22, 2170–2181 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolas E., et al. , Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 14, 372–380 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Cai R. L., Yan-Neale Y., Cueto M. A., Xu H., Cohen D., HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275, 27909–27916 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Nag R., Kyriss M., Smerdon J. W., Wyrick J. J., Smerdon M. J., A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast. Nucleic Acids Res. 38, 1450–1460 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyrick J. J., Kyriss M. N., Davis W. B., Ascending the nucleosome face: Recognition and function of structured domains in the histone H2A-H2B dimer. Biochim. Biophys. Acta 1819, 892–901 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T., The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 23, 9178–9188 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saldivar J. C., Cortez D., Cimprich K. A., The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschiersch B., et al. , The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13, 3822–3831 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rea S., et al. , Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I., Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Couch F. B., et al. , ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kai M., Boddy M. N., Russell P., Wang T. S., Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 19, 919–932 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyabe I., Morishita T., Shinagawa H., Carr A. M., Schizosaccharomyces pombe Cds1Chk2 regulates homologous recombination at stalled replication forks through the phosphorylation of recombination protein Rad60. J. Cell Sci. 122, 3638–3643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassin V. M., Anantha R. W., Sokolova E., Kanner S., Borowiec J. A., Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 122, 4070–4080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng J. C., Karpen G. H., H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9, 25–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi X., et al. , Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol. Biol. Cell 20, 983–994 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubarry M., Loïodice I., Chen C. L., Thermes C., Taddei A., Tight protein-DNA interactions favor gene silencing. Genes Dev. 25, 1365–1370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller A. M., Nasmyth K. A., Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312, 247–251 (1984). [DOI] [PubMed] [Google Scholar]
- 42.Kunkel T. A., Erie D. A., Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall I. M., et al. , Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Volpe T. A., et al. , Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Taverna S. D., Coyne R. S., Allis C. D., Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110, 701–711 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Mochizuki K., Fine N. A., Fujisawa T., Gorovsky M. A., Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110, 689–699 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Kanoh J., Sadaie M., Urano T., Ishikawa F., Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 15, 1808–1819 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Grewal S. I., Klar A. J., A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146, 1221–1238 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.