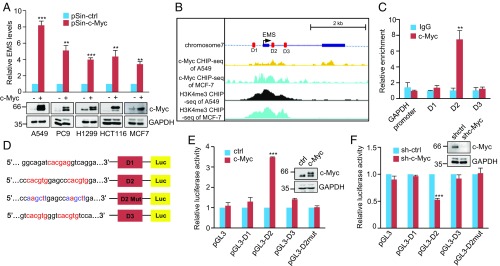
Fig. 2.
EMS is a direct c-Myc transcriptional target. (A) Indicated cells were infected with lentiviruses expressing control (pSin-ctrl) or c-Myc (pSin-c-Myc) protein. Forty-eight hours later, total RNA was analyzed by real-time RT-PCR. Data shown are mean ± SD (n = 3). **P < 0.01; ***P < 0.001. (B) Encode ChIP-seq data for c-Myc and H3K4me3 are displayed in the UCSC Genome Browser illustrations. Black boxes indicate 2 exons of EMS. D1, D2, and D3 represent 3 putative c-Myc binding sites predicted by the JASPAR database. (C) Lysates from A549 cells were subjected to ChIP assay using anti–c-Myc antibody or an isotype-matched control immunoglobulin G (IgG). ChIP products were amplified by real-time PCR. Data shown are mean ± SD (n = 3). **P < 0.01. (D) Shown are the pGL3-based wild-type and mutant reporter constructs used for luciferase (Luc) assay. (E) A549 cells were cotransfected with control vector (ctrl), Flag-c-Myc, or together with the reporter constructs in the indicated combination. Twenty-four hours after transfection, reporter activity was measured and plotted after normalizing with respect to Renilla luciferase activity. Data shown are mean ± SD (n = 3). ***P < 0.001. (F) A549 cells transduced with lentiviruses expressing control shRNA (sh-Ctrl) or c-Myc shRNA (sh-c-Myc) were cotransfected with the indicated reporter constructs. Reporter activity was then measured and plotted after normalizing with respect to Renilla luciferase activity. Data shown are mean ± SD (n = 3). ***P < 0.001.