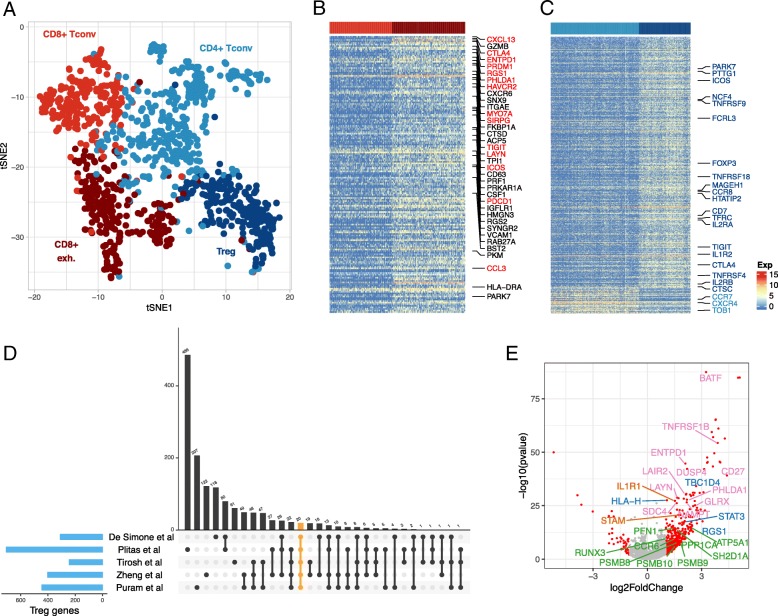
Fig. 2.
Deconvolution of T cell subtypes. a 2D t-sne projection of T cells. T cell subtypes identified by clustering analysis are annotated and marked by color codes. b Heatmap of genes significantly expressed in exhausted CD8+ T cells comparing to conventional CD8+ T cells (adjusted p-value <0.05, log2fold-change > 1). Genes also reported by a previous study are labeled on left, of which the known exhaustion markers are labeled in red text. Cell types are indicated by the colored bar at top. c Heatmap of genes differentially expressed in Tregs comparing with conventional CD4+ T cells (adjusted p-value < 0.05, |log2fold-change| > 1). Selective Treg genes are labeled in dark blue and known markers for conventional CD4+ T cells are labeled in light blue. d Comparing Treg genes identified in (c) with Treg genes reported by previous four studies. The combination matrix at the bottom indicates all intersections of any of the five studies. If a study is participating in an interaction, the corresponding matrix cell is filled with black. All studies participating in the same interaction are linked by lines. The bars above the combination matrix encode the size of each intersection. The 20 Treg genes shared by all five studies are highlighted in orange and also labeled in (c). e Volcano plot of genes differentially expressed in Tregs vs. conventional CD4+ T cells. Unique genes found by this study are labeled in green. Those identified once (blue), twice (red), and three times (pink) previously are also labeled