Abstract
Background
This study aimed to investigate the association between the rs2975760 and rs3792267 single nucleotide polymorphisms (SNPs) of the calpain 10 (CAPN10) gene and gestational diabetes mellitus.
Material/Methods
The study included 138 patients with gestational diabetes mellitus and 152 healthy pregnant women. Venous blood was separated, and the DNA was extracted. The rs2975760 and rs3792267SNP polymorphisms of CAPN10 were detected using polymerase chain reaction (PCR). The frequencies of different genotypes in patients with gestational diabetes mellitus and healthy pregnant women were determined, and the relationship between different SNP genotypes and the risk of gestational diabetes mellitus was analyzed.
Results
There were no significant differences in the frequencies of the TT, CT and CC genotypes of rs2975760 and the frequencies of the GG, AG and AA genotypes of rs3792267 between the women with gestational diabetes and the controls. Expression of rs2975760 and rs3792267 were not associated with the risk of gestational diabetes in the dominant model, recessive model, and additive model. However, grade B and grade D diabetes in the CC and TC genotypes of rs2975760 were significantly different from those in the TT genotype (P<0.05). Grade B and grade D diabetes in the AA and AG genotypes of rs3792267 were significantly different compared with those in the GG genotype (P<0.05), and allele A was significantly increased compared with allele G (P<0.05).
Conclusions
The rs2975760 and rs3792267 SNP polymorphisms of CAPN10 showed no significant association with the incidence of gestational diabetes mellitus and only a mild association with the severity.
MeSH Keywords: Calpain; Diabetes, Gestational; Genotype
Background
Gestational diabetes mellitus is the condition of increased blood glucose above the normal level before pregnancy or impaired glucose tolerance in women during pregnancy [1,2]. Gestational diabetes mellitus is harmful to the pregnant woman and the fetus and increases the risk of pregnancy-induced hypertension, ketoacidosis, excessive amniotic fluid, and abortion [3,4]. The probability of dystocia, birth canal injury and cesarean section are also increased [3,4]. In gestational diabetes, the prevalence of fetal macrosomia is as high as 25–42%, which inhibits the development of the embryo and leads to reduced fetal development in early pregnancy [5,6]. Gestational diabetes mellitus is a cause of fetal malformation, fetal abortion, premature birth, and fetal death [5,6]. However, the causes of gestational diabetes mellitus remain unclear. In most cases, diabetes mellitus only appears after pregnancy, and is related to the environment, eating habits and hormones, and genetic factors in different populations [7, 8].
Expression of the calpain 10 gene, CAPN10, has been shown to be associated with type 2 diabetes mellitus, and it is closely associated with blood glucose and insulin secretion. The mutation of base pairs in the CAPN10 gene results in amino acid changes that increase in blood glucose [9–11].
This study aimed to investigate the association between the rs2975760 and rs3792267 single nucleotide polymorphisms (SNPs) of CAPN10 and gestational diabetes mellitus.
Material and Methods
Patient data
The study included 138 women with gestational diabetes mellitus who were treated in our hospital from June 2015 to June 2018 and 152 healthy pregnant women with normal blood glucose. The study inclusion criteria included a diagnosis of gestational diabetes mellitus, a single pregnancy from natural conception, a maternal examination performed at our hospital, and agreement to participate in the study. Study exclusion criteria included pregnant women with liver or kidney disease, diabetes, hypertension, or other medical diseases before pregnancy, endocrine diseases that included Cushing’s syndrome, hypothyroidism, and hyperthyroidism, and refusal to participate in the study. All women signed informed consent, and the study was approved by the Ethics Committee of Jining No. 1 People’s Hospital.
Grading of patients with gestational diabetes mellitus
According to age, the course and the presence of vascular complications, patients with gestational diabetes mellitus were graded using the White classification for maternal and fetal risk in gestational diabetes mellitus. The following grading categories were used: Grade A, after dietary control, a fasting blood glucose of <5.8 mmol/L and a 2-hour postprandial blood glucose <6.7 mmol/L; Grade B, diabetes mellitus occurring >20 years with a course of <10 years; Grade C, and age of onset of diabetes mellitus that ranged from 10–19 years, or a course of disease that ranged from 10–19 years; Grade D, onset of diabetes mellitus of <10 years, or duration >20 years, or combined diabetic retinopathy.
Extraction of the genomic DNA
All subjects provided 5 mL of whole blood that was collected during the regular prenatal examination at 24–28 weeks’ gestation. The genomic DNA was extracted from the blood using Omega Mag-Bind DNA Kit (OmegaBiotek, Doraville, GA, USA), and the concentration and purity of DNAs were measured using a NanoDrop spectrophotometer (Thermofisher Scientific, Waltham, MA, USA). DNA was stored at −20°C.
Single-nucleotide polymorphism (SNP) genotyping using polymerase chain reaction (PCR)
The primer sequence at the SNP site and its TaqMan probe sequence were designed using Oligo 6.0 primer design software (www.oligo.net/) (Table 1). Primers were synthesized by Beijing Genomics Institute (BGI, Beijing, China). Initially, 1 μL of DNA solution and 1.2 μL of primer solution, including 0.4 μL of forward and reverse primers and 0.4 μL of probe primers, were added to the pre-configured 17.8 μL of TransStart Probe quantitative PCR (qPCR) SuperMix (Beijing TransGene Biotech Co., Ltd, Beijing, China) and evenly mixed by slight shaking and placed in a CFX96 fluorescence qPCR instrument (Bio-Rad, Hercules, CA, USA). The PCR reaction conditions were: 94°C for 3 min, 94°C for 10 sm and 60°C for 15 s for a total of 40 cycles. The instrument software generated the experimental results. Each sample was analyzed in triplicate. Diethyl pyrocarbonate (DEPC) water was used as the negative control, and a positive plasmid containing the sequence was used as the positive control (Shanghai Sangon Biotech Co., Ltd., Shanghai, China). The C, T, A, G alleles were only analyzed in relation to the grades of gestational diabetes mellitus due to a limitation of funding and time.
Table 1.
Primer sequences.
| SNP | Primer sequence | Probe sequence |
|---|---|---|
| rs2975760 | Forward: 5′-CCCTGCTCCTGTGCCCACAC-3′ | HEX: 5′-CGCTTGCTGTGAAGTAAGGCGTT-3′ |
| Reverse: 5′-AAATCGTCCAACCGCTGCCT-3′ | FAM: 5′-CGCTTGCTGCGAAGTAAGGCGTT -3′ | |
| rs3792267 | Forward: 5′-AGCGGAGGACGGGACGAGGA-3′ | HEX: 5′-AGGCGTTTGAAGGTGAGGCTA-3′ |
| Reverse: 5′-TCGTCCAACCGCTGCCTCAAC-3′ | FAM: 5′-AGGCATTTGAAGGTGAGGCTA -3′ |
Statistical analysis
Data were analyzed using SPSS version 19.0 software (IBM, Armonk, NY, USA). The difference in the distribution of genotypes between the study group and the control group was analyzed using the chi-squared (χ2) test, and the relationship between each genotype and the risk of gestational diabetes mellitus was determined using logistic regression analysis. A P-value <0.05 was considered to be statistically significant.
Results
Clinical and demographic data
There were no significant differences in age, living habits, body mass index (BMI), and parity between the study group and the control group (P>0.05) (Table 2). According to White grading, there were 51 women with Grade A, 39 women with Grade B, 26 women with Grade C, and 22 women with Grade D gestational diabetes mellitus.
Table 2.
Clinical and demographic data of women in the study group and control group.
| Index | Study group (n=138) | Control group (n=152) | P-value |
|---|---|---|---|
| Age (years) | 28.2±9.3 | 27.5±8.3 | 0.423 |
| Smoking | |||
| Yes | 32 (23.19%) | 45 (29.61%) | 0.303 |
| No | 106 (76.81%) | 107 (70.39%) | |
| Drinking | |||
| Yes | 67 (48.55%) | 74 (48.68%) | 1 |
| No | 71 (51.45%) | 78 (51.32%) | |
| BMI | 24.3±4.7 | 23.7±4.1 | 0.401 |
| Parity | |||
| 0 | 75 (54.35%) | 83 (54.61%) | 0.587 |
| 1 | 34 (24.64%) | 41 (26.97%) | |
| ≥2 | 29 (21.01%) | 28 (18.42%) | |
TaqMan polymerase chain reaction (PCR) genotyping results of the two single nucleotide polymorphisms (SNPs) of CAPN10, rs2975760, and rs3792267
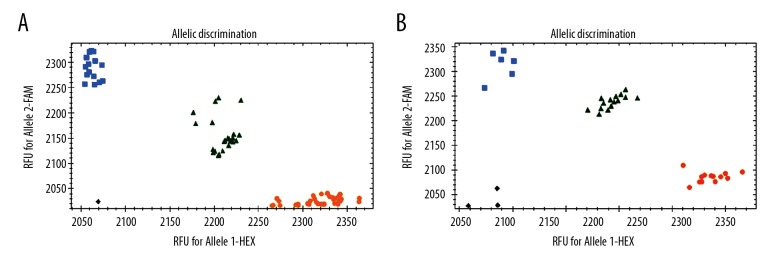
The 138 patients with gestational diabetes mellitus and 152 healthy pregnant women all had good genotyping results (Figure 1). The distribution conformed to the Hardy-Weinberg equilibrium: P (rs2975760)=0.31, and P (rs3792267)=0.58.
Figure 1.
Genotyping results of the rs2975760 and rs3792267 single nucleotide polymorphisms (SNPs) of CAPN10. (A) Genotyping results of rs2975760. (B) Genotyping results of rs3792267. At rs2975760, HEX is marked as TT, FAM is marked as CC, and the middle green part is CT. At rs3792267, HEX is labeled as GG, FAM is labeled as AA, and the middle green part is AG.
Association between CAPN10 SNP polymorphism and the occurrence of gestational diabetes mellitus
Differences in the frequencies of TT, CT, and CC genotypes of rs2975760 between the study group and the control group were not significant (P>0.05). The frequencies of GG, AG, and AA genotypes of rs3792267 were not significant between the study group and the control group (P>0.05) (Table 3).
Table 3.
Association between single nucleotide polymorphisms (SNPs) of the calpain 10 (CAPN10) gene and gestational diabetes mellitus.
| Genotype | Study group | Control group | OR (95% CI) | P-value | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| rs2975760 | ||||||
| TT | 53 | 38.41 | 65 | 42.76 | 1 | 0.531 |
| CT | 68 | 49.28 | 79 | 51.97 | 1.056 (0.923–1.332) | 0.856 |
| CC | 17 | 12.31 | 8 | 5.27 | 2.606 (2.232–3.014) | 0.078 |
| rs3792267 | ||||||
| GG | 95 | 68.84 | 113 | 74.34 | 1 | 0.388 |
| AG | 35 | 25.36 | 36 | 23.68 | 1.156 (0.822–1.439) | 0.783 |
| AA | 8 | 5.80 | 3 | 1.97 | 2.743 (2.213–3.321) | 0.161 |
Analysis results of the risk of gestational diabetes mellitus through different models
There was no risk of gestational diabetes mellitus in the dominant model, the recessive model, and the additive model of the rs2975760 and rs3792267 SNP polymorphisms of CAPN10 (P>0.05) (Table 4).
Table 4.
Risk of gestational diabetes associated with the rs2975760 and rs3792267 single nucleotide polymorphisms (SNPs) of CAPN10.
| Model | Genotype | Study group (n=138) | Control group (n=152) | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| rs2975760 | |||||
| Dominant model | TT | 53 (38.41) | 65 (42.76) | 1 | 0.531 |
| CT+CC | 85 (61.59) | 87 (57.24) | 1.198 (0.757–1.416) | ||
| Recessive model | TT+TC | 121 (87.69) | 144 (94.73) | 1 | 0.079 |
| CC | 17 (12.31) | 8 (5.27) | 2.529 (1.825–4.044) | ||
| Additive model | TT | 53 (38.41) | 65 (42.76) | 1 | 0.084 |
| CC | 17 (12.31) | 8 (5.27) | 2.606 (1.923–3.532) | ||
| rs3792267 | |||||
| Dominant model | GG | 95 (68.84) | 113 (74.34) | 1 | 0.388 |
| AG+AA | 43 (31.16) | 87 (25.66) | 1.198 (0.757–1.416) | ||
| Recessive model | GG+AG | 130 (94.2) | 149 (98.03) | 1 | 0.161 |
| AA | 8 (5.8) | 3 (1.97) | 2.529 (1.825–4.044) | ||
| Additive model | GG | 95 (68.84) | 113 (74.34) | 1 | 0.149 |
| AA | 8 (5.8) | 3 (1.97) | 2.606 (1.923–3.532) | ||
Relationship between gene polymorphism and white grading in patients with gestational diabetes mellitus patients
Grade B and grade D cases of diabetes mellitus in the CC and TC genotypes of rs2975760 were significantly different from those in TT genotype (P<0.05), and allele C was higher than allele T (P<0.05) (Table 4). Grade B and grade D cases of diabetes mellitus in the AA and AG genotypes of rs3792267 were significantly different from those in GG genotype (P<0.05), and allele A was higher than allele G (P<0.05) (Table 5).
Table 5.
Association between the grade of gestational diabetes mellitus and the rs2975760 and rs3792267 single nucleotide polymorphisms (SNPs) of CAPN10.
| Grade | rs2975760 | rs3792267 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TT (n=53) | TC (n=68) | CC (n=17) | T (n=174) | C (n=102) | GG (n=95) | AG (n=35) | AA (n=8) | G (n=225) | A (n=51) | |
| A | 4 (7.55) | 3 (4.41) | 1 (5.88) | 11 (6.32) | 5 (4.9) | 6 (6.32) | 3 (8.57) | 1 (12.5) | 19 (8.44) | 5 (9.8) |
| B | 24 (45.28) | 21 (30.88)* | 5 (29.41)* | 69 (39.66) | 31 (30.39) | 48 (50.53) | 10 (28.57)* | 2 (25)* | 106 (47.11) | 14 (27.45) |
| C | 22 (41.51) | 30 (44.12) | 7 (41.18) | 74 (42.53) | 44 (43.14) | 37 (38.95) | 19 (54.29) | 3 (37.5) | 83 (36.89) | 25 (49.02) |
| D | 3 (5.66) | 14 (20.59)* | 4 (23.53)* | 20 (11.49) | 22 (22.57)# | 4 (4.21) | 3 (8.57) | 2 (25)* | 11 (4.89) | 7 (13.73)# |
P<0.05 vs. TT, and
P<0.05 vs. T.
Discussion
The control of blood glucose levels in gestational diabetes mellitus uses diet, drugs, and lifestyle changes. Early detection and control of gestational diabetes mellitus improves the health of both the pregnant woman and the fetus [12,13]. Therefore, early detection and prevention of gestational diabetes mellitus will benefit from the identification of susceptible women. The calpain 10 (CAPN10) gene has a role in the control of blood glucose concentrations and structural changes in the calpain 10 protein encoded by this gene affect the increase in blood glucose [14,15]. Gene polymorphisms are an important manifestation of genetic changes and genetic differences between people [16]. Previous studies have shown that gene polymorphisms of CAPN10 are correlated with type 2 diabetes mellitus in China, and the waist-hip ratio and body mass index of patients with the GG genotype of SNP43 were relatively high [17]. Other studies have shown that the distribution frequency of single nucleotide polymorphisms (SNPs) of CAPN10 are different between different ethnicities and populations, and so identification of the same change in genotype cannot be used to identify the susceptibility of Europeans and Asians to gestational diabetes mellitus [18,19].
Based on the correlation between SNP polymorphisms of CAPN10 and type 2 diabetes mellitus, in this study on gestational diabetes mellitus the findings showed that the proportions of mutant-type homozygotes of two typical SNPs (rs2975760 and rs3792267) of CAPN10 were less than those of wild-type homozygotes. Although the number of mutations in the study group was higher than that in the control group, the change in the SNP polymorphisms had no significant association with gestational diabetes mellitus, even though there were more mutant heterozygotes. If heterozygotes were affected by the external environment or metabolic conditions, the increased probability of mutant-type homozygotes would be increased. Therefore, pregnant women with mutant genotypes of CAPN10 may be identified as having an increased risk of gestational diabetes mellitus, and the impact of base mutations should be appreciated. The findings from previous studies have indicated that the base mutations may reduce the activity of the encoded non-lysosomal cysteine proteases and the ability to degrade blood glucose [20].
In this study, SNP polymorphisms of CAPN10 and different White grades of gestational diabetes mellitus were analyzed. The results showed that genotypes CC and TC of rs2975760 after mutation were significantly different in women with grade B and grade D gestational diabetes mellitus when compared with genotype TT, and allele C was higher than allele T. The AA and AG genotypes of rs3792267 were significantly different in women with grade B and grade D gestational diabetes mellitus compared with genotypes GG, and allele A was higher than allele G. The odds ratio (OR) was 2.6 in both the rs2975760 and rs3792267 polymorphisms. However, further studies are needed to determine whether there are correlations between these two polymorphisms.
The findings from this study indicate that the mutant CAPN10 gene had an effect on the grade of gestational diabetes mellitus in women, and the proportion of women with mutations in the higher grades of the disease was relatively large. These findings may have clinical implications for the identification of women who are at increased risk of gestational diabetes mellitus. However, in the present study, we were only able to evaluate alleles C, T, A and G in relation to the grades of gestational diabetes mellitus, due to the limitations of funding and time. Also, the sample size of this study was small, and future large-scale studies are needed to validate the findings.
Conclusions
The rs2975760 and rs3792267 single nucleotide polymorphisms (SNPs) of the calpain 10 (CAPN10) gene showed no significant association with the incidence of gestational diabetes mellitus and only a mild association with the severity.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Macri F, Pitocco D, di Pasquo E, et al. Gestational weight gain as an independent risk factor for adverse pregnancy outcomes in women with gestational diabetes. Eur Rev Med Pharmacol Sci. 2018;22:4403–10. doi: 10.26355/eurrev_201807_15490. [DOI] [PubMed] [Google Scholar]
- 2.Moyce BL, Dolinsky VW. Maternal beta-cell adaptations in pregnancy and placental signalling: Implications for gestational diabetes. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19113467. pii: E3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand JS, West J, Tuffnell D, et al. Gestational diabetes and ultrasound-assessed fetal growth in South Asian and White European women: Findings from a prospective pregnancy cohort. BMC Med. 2018;16:203. doi: 10.1186/s12916-018-1191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellos I, Fitrou G, Pergialiotis V, et al. Serum levels of adipokines in gestational diabetes: A systematic review. J Endocrinol Invest. 2018 doi: 10.1007/s40618-018-0973-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Goveia P, Canon-Montanez W, Santos DP, et al. Lifestyle intervention for the prevention of diabetes in women with previous gestational diabetes mellitus: A systematic review and meta-analysis. Front Endocrinol. 2018;9:583. doi: 10.3389/fendo.2018.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carolan-Olah M, Sayakhot P. A randomized controlled trial of a web-based education intervention for women with gestational diabetes mellitus. Midwifery. 2019;68:39–47. doi: 10.1016/j.midw.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Cremona A, O’Gorman C, Cotter A, et al. Effect of exercise modality on markers of insulin sensitivity and blood glucose control in pregnancies complicated with gestational diabetes mellitus: A systematic review. Obes Sci Pract. 2018;4:455–67. doi: 10.1002/osp4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davenport MH, Ruchat SM, Poitras VJ, et al. Prenatal exercise for the prevention of gestational diabetes mellitus and hypertensive disorders of pregnancy: A systematic review and meta-analysis. Br J Sports Med. 2018;52:1367–75. doi: 10.1136/bjsports-2018-099355. [DOI] [PubMed] [Google Scholar]
- 9.Wu K, Cai Y. The SNP43 (G/A) polymorphism in CAPN10 gene confers an increased risk of cognitive impairment in cerebral small vessel disease. J Clin Lab Anal. 2018;32:e22615. doi: 10.1002/jcla.22615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro-Martinez AG, Sanchez-Corona J, Vazquez-Vargas AP, et al. Association analysis of calpain 10 gene variants/haplotypes with gestational diabetes mellitus among Mexican women. Cell Mol Biol. 2018;64:81–86. doi: 10.14715/cmb/2018.64.3.13. [DOI] [PubMed] [Google Scholar]
- 11.Thangavelu M, Godla UR, Paul SF, Maddaly R. Single-nucleotide polymorphism of INS, INSR, IRS1, IRS2, PPAR-G and CAPN10 genes in the pathogenesis of polycystic ovary syndrome. J Genet. 2017;96:87–96. doi: 10.1007/s12041-017-0749-z. [DOI] [PubMed] [Google Scholar]
- 12.Mak JKL, Lee AH, Pham NM, et al. Gestational diabetes and postnatal depressive symptoms: A prospective cohort study in Western China. Women Birth. :2018. doi: 10.1016/j.wombi.2018.08.171. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Lekva T, Godang K, Michelsen AE, et al. Prediction of gestational diabetes mellitus and pre-diabetes 5 years postpartum using 75 g oral glucose tolerance test at 14–16 weeks’ gestation. Sci Rep. 2018;8:13392. doi: 10.1038/s41598-018-31614-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plengvidhya N, Chanprasert K, Tangjittipokin W, et al. Detection of CAPN10 copy number variation in Thai patients with type 2 diabetes by denaturing high-performance liquid chromatography and real-time quantitative polymerase chain reaction. J Diabetes Investig. 2015;6:632–39. doi: 10.1111/jdi.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui J, Xu X, Yin S, et al. Meta-analysis of the association between four CAPN10 gene variants and gestational diabetes mellitus. Arch Gynecol Obstet. 2016;294:447–53. doi: 10.1007/s00404-016-4140-8. [DOI] [PubMed] [Google Scholar]
- 16.Hou Z, Li M, Cao Y. TCF7L2, CAPN10 polymorphisms are associated with gestational diabetes mellitus (GDM) risks: A meta-analysis. Gynecol Endocrinol. 2017;33:399–404. doi: 10.1080/09513590.2017.1290066. [DOI] [PubMed] [Google Scholar]
- 17.Zhao F, Mamatyusupu D, Wang Y, et al. The Uyghur population and genetic susceptibility to type 2 diabetes: Potential role for variants in CAPN10, APM1 and FUT6 genes. J Cell Mol Med. 2016;20:2138–47. doi: 10.1111/jcmm.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayramci NS, Acik L, Kalkan C, Yetkin I. Investigation of glucocorticoid receptor and calpain-10 gene polymorphisms in Turkish patients with type 2 diabetes mellitus. Turk J Med Sci. 2017;47:1568–75. doi: 10.3906/sag-1701-174. [DOI] [PubMed] [Google Scholar]
- 19.Reddy BM, Kommoju UJ, Dasgupta S, Rayabarapu P. Association of type 2 diabetes mellitus genes in polycystic ovary syndrome aetiology among women from southern India. Indian J Med Res. 2016;144:400–8. doi: 10.4103/0971-5916.198678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashyap S, Kumar S, Agarwal V, et al. Gene expression profiling of coronary artery disease and its relation with different severities. J Genet. 2018;97:853–67. [PubMed] [Google Scholar]