Abstract
Helicases are enzymes that move, manage, and manipulate nucleic acids. They can be subdivided into six super families and are required for all aspects of nucleic acid metabolism. In general, all helicases function by converting the chemical energy stored in the bond between the gamma and beta phosphates of adenosine triphosphate into mechanical work, which results in the unidiresctional movement of the helicase protein along one strand of a nucleic acid. The results of this translocation activity can range from separation of strands within duplex nucleic acids to the physical remodeling or removal of nucleoprotein complexes. In this review, we focus on describing key helicases from the model organism Saccharomyces cerevisiae that contribute to the regulation of homologous recombination, which is an essential DNA repair pathway for fixing damaged chromosomes.
Keywords: homologous recombination, helicase, Srs2, Sgs1, Rad54
INTRODUCTION
There are ~95 helicases encoded within the human genome, and these proteins participate in nearly all aspects of nucleic acid metabolism, including maintenance of genome integrity (11, 18–20, 39, 98). Helicases utilize the energy derived from adenosine triphosphate (ATP) hydrolysis to translocate along one nucleic acid strand while displacing the other strand (77, 109, 120). They can also remodel or disrupt other nucleoprotein complexes (11, 18–20, 39, 98).
Helicases share a related core domain composed of two RecA-like folds, which couple binding and hydrolysis of ATP to conformational changes that propel the helicase along a nucleic acid (77, 109, 120, 121). Although many helicases share basic properties, their differences dictate biological functions. Strand polarity and substrate binding specificity both influence biological function. For instance, some helicases move in a 3′→5′ direction, whereas others move 5′→3′; some helicases bind single-stranded DNA (ssDNA), and others bind double-stranded DNA (dsDNA) or ssDNA/dsDNA junctions (77, 109, 120, 121). Many helicases participate in RNA metabolism and thus bind preferentially to RNA (77, 109, 120, 121). Given their range of functions, helicases provide an important example of how evolution has used a single protein domain (i.e., the RecA-like domain) to produce a range of enzymes capable of diverse biological processes.
Here, we describe how helicases from Saccharomyces cerevisiae (baker’s yeast) regulate homologous recombination (HR). We focus on S. cerevisiae helicases because there is a wealth of genetic, biochemical, and biophysical information describing them. Examples of these proteins include Sgs1, Mph1, Srs2, Rad54, and Rdh54, all of which are broadly conserved (11, 17–19, 29, 56, 79, 98, 137, 142). We discuss how these proteins participate in HR and highlight future questions that will need to be addressed to more fully appreciate their functions.
HOMOLOGOUS RECOMBINATION
Double-strand breaks (DSBs) can cause cell death, cancer, or severe genetic disorders (65, 87, 107). DSBs can arise from exogenous factors, such as ionizing radiation, or endogenous causes, such as spontaneous DNA replication errors (65, 87, 107). Programmed DSBs are essential to ensure proper chromosome segregation during meiosis, part of the eukaryotic life cycle required for gamete production (58, 66, 94, 148). HR is an important pathway used for repairing DSBs (60, 101, 127). Its importance is further underscored by the finding that null mutations in HR genes are often embryonic lethal in mice (33, 42, 69, 74, 86, 104, 107, 118, 119, 122, 131), the hypersensitivity to DNA-damaging agents and replication stress observed in many HR-deficient cells (45, 78, 122), the association of cancers and cancer syndromes in patients with HR mutations (6, 16, 44, 116), and the links between DSB repair and age-related decline in human health (138). The central feature of HR is its use of an undamaged copy of the broken DNA as a template to guide repair (Figure 1a) (60, 65, 87, 101, 107, 127).
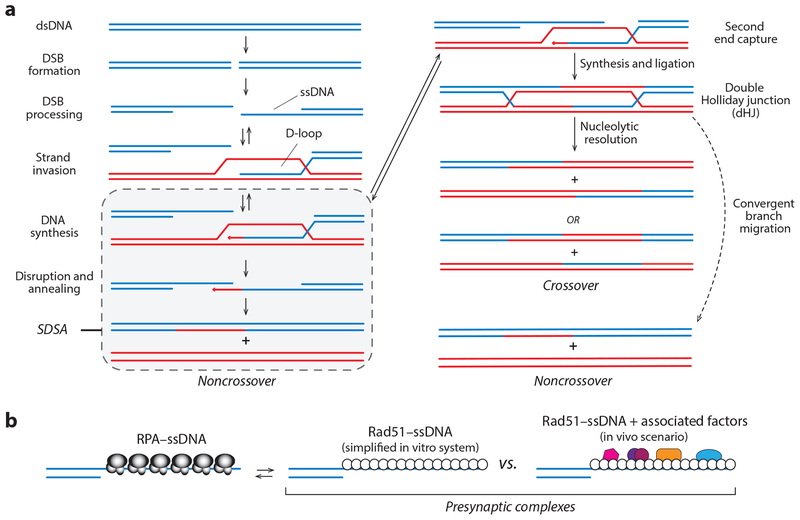
Figure 1.
(a) HR is initiated by 5′→3′ resection of the DNA ends. The ssDNA overhangs are paired with a homologous dsDNA that is used as atemplate for repair. In mitotic cells, these intermediates are normally channeled through the SDSA pathway. Alternatively, the second ssDNA can also pair with the dsDNA, leading to formation of a dHJ, which can be resolved through convergent branch migration to yield noncrossovers or cleaved by nucleases to yield crossovers. (b) Early nucleoprotein complexes in HR. The 3′ ssDNA overhangs are first bound by RPA, which is then displaced by Rad51 to form the presynaptic complex. In vitro, Rad51–ssDNA can serve as a minimal system for promoting strand invasion. In vivo, the presynaptic complex comprises Rad51 plus numerous cofactors (reviewed in 60, 67, 87, 101, 127). Abbreviations: dHJ, double Holliday junction; DSB, double-strand break; dsDNA, double-stranded DNA; HR, homologous recombination; RPA, replication protein A; SDSA, synthesis-dependent strand annealing; ssDNA, single-stranded DNA.
HR begins with formation of a DSB (Figure 1) (60, 101, 127). Broken DNA ends are resected in the 5′→3′ direction to yield 3′ssDNA overhangs (60, 101, 127). These overhangs are bound by the heterotrimeric protein complex RPA (replication protein A), which protects the ssDNA and removes the secondary structure (32, 139). Rad52 stimulates the replacement of RPA with Rad51 (60, 101, 127). Rad51 is the only recombinase present in mitosis, whereas both Rad51 and the second recombinase called Dmc1 are present during meiosis (21, 94). These recombinases are ATP-dependent DNA-binding proteins that form extended helical filaments, referred to as presynaptic complexes (67, 89). The presynaptic complex performs a homology search to locate a donor dsDNA with sequence homology to the Rad51-bound ssDNA. The presynaptic complex then catalyzes strand invasion, which results in the formation of a D-loop where the presynaptic ssDNA is paired with the homologous donor. The 3′ end of the invading ssDNA is used as a primer for DNA synthesis, allowing information from the donor template to be used to replace information that has been lost from the damaged DNA (60, 101, 127).
Repair can be complete through either the SDSA (synthesis-dependent strand annealing) or DSBR (DSB repair) pathways (reviewed in 60, 67, 87, 101, 127) (Figure 1a). In mitotic cells, HR intermediates are channeled through SDSA, which minimizes the potential for chromosomal rearrangements and results in noncrossover recombination products. During SDSA, the second DSB end does not pair with the homologous dsDNA but is instead reannealed to the newly synthesized ssDNA present on the other DSB end (Figure 1a). Alternatively, the second DSB end can pair with the dsDNA template, leading to formation of a double Holliday junction (dHJ), which can be resolved through either convergent branch migration to yield noncrossover recombination products or the action of nucleases to yield crossovers (Figure 1a). Crossovers are essential for proper chromosome segregation during meiosis (21, 58, 66, 94, 148).
MECHANISMS OF HELICASE TRANSLOCATION
Helicases couple both ATP binding and hydrolysis to movement along nucleic acids. Detailed understanding of helicase translocation comes from studies of the bacterial superfamily 1 helicases UvrD and PcrA (72, 123, 135, 143), which suggest the existence of an inchworm-like mechanism (Figure 2) (reviewed in 143). This mechanism involves two central features: (a) ATP hydrolysis–dependent structural transitions involving the relative motions of two protein domains, which propel the enzymes forward, and (b) at least two points of contact with the nucleic acids that undergo coupled changes in nucleic acid–binding affinity, which allow for cycles of tight and loose binding interactions that enable the proteins to move forward along a nucleic acid track (Figure 2) (143).
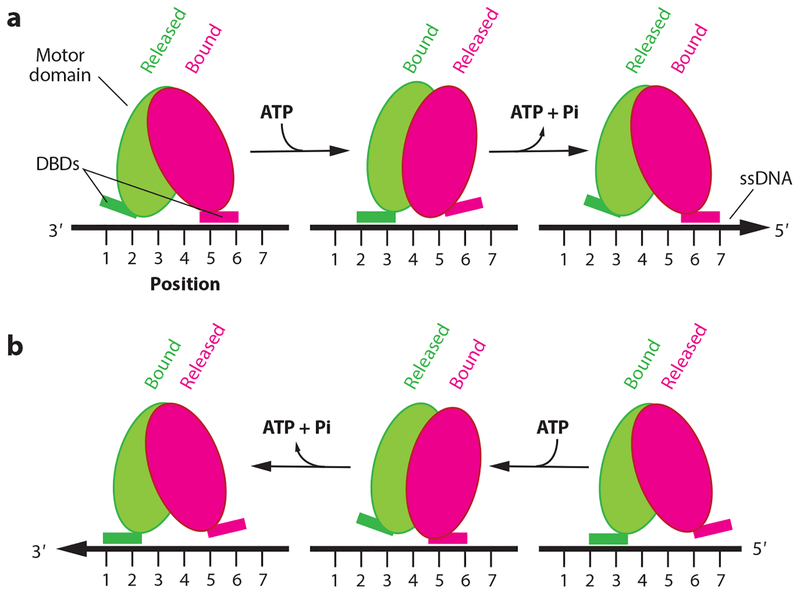
Figure 2.
Inchworm mechanism. (a) Tandem RecA-like domains composing the motor domain of the helicase (magenta and green ovals). The motor domain interacts with the ssDNA track through DBDs, which alternate between tightly (bound) and weakly (released) bound states. ATP binding is also coupled to rotation of the RecA domains and changes in the ssDNA affinity of the DBDs, thus propelling the helicase in the 3′→5′ direction. (b) The same mechanism can propel the helicase in the opposite direction (5′→3′) if tightly (bound) and weakly (released) bound states are reversed with respect to the ATP binding/hydrolysis cycle. Figure adapted from Reference 143. Abbreviations: DBD, DNA-binding domain; ssDNA, single-stranded DNA.
These principles are supported by detailed atomic-level structural studies of UvrD, which have revealed that the two RecA-like domains that make up the central core of each protein undergo a large rotation upon ATP binding, and the rotation is reversed upon ATP hydrolysis and ADP/Pi release (143). The domain rotations are converted to linear movement through DNA contact points, and changes in their relative DNA-binding affinities are coupled to the motions of the RecA-like domains (143). Given the inchworm model, the distance that a helicase moves during one round of ATP binding, hydrolysis, and release is dictated by the magnitude of the protein domain motions, and translocation is determined by how the loose- and tight-binding states of the nucleic acid–interaction domains are coupled to the ATP hydrolysis cycle (Figure 2) (143). It is likely that these general mechanistic principles for converting the chemical energy of ATP into motion along a nucleic acid are shared among many helicases.
HELICASE CONTROL OF HOMOLOGOUS RECOMBINATION
The major steps in HR include DNA end resection, assembly of the presynaptic complex, homology search, DNA-strand invasion, and HJ formation and resolution (Figure 1). Each of these steps requires the actions of helicases, and the main helicases in S. cerevisiae include Sgs1, Srs2, Mph1, Rad54, and Rdh54 (67, 127). In subsequent sections, we describe the participation of these helicases in HR.
Srs2 Disrupts Rad51 Filaments
Srs2 is a UvrD-like superfamily 1 helicase and is a negative regulator of Rad51 (reviewed in 80, 98). The SRS2 gene was identified in a genetic screen that suppressed the sensitivity of rad6 mutants to DNA-damaging agents (1) and was later shown to harbor canonical helicase motifs (113). Srs2 is a 134-kDa protein (1,174 amino acids) that has ssDNA-dependent ATP hydrolysis activity (kcat ~ 300 s−1) and is capable of unwinding dsDNA structures (112, 132). Srs2 also contains a Rad51-interaction domain near its C terminus (80, 98). SRS2 mutants typically exhibit a hyper-recombination phenotype, suggesting that Srs2 normally restrains HR (43, 59, 79, 100, 103). Indeed, biochemical studies have shown that Srs2 translocates in the 3′→5′ direction along ssDNA and in doing so strips Rad51 from ssDNA (5, 68, 134). This activity has earned Srs2 the moniker antirecombinase and explains how Srs2 affects HR (Figure 3) (17, 18).
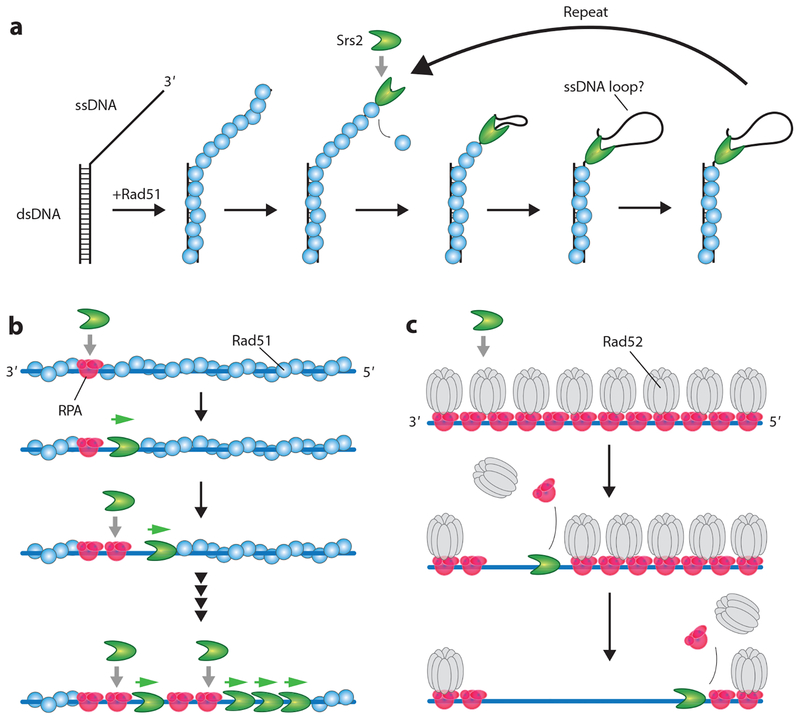
Figure 3.
Srs2 disruption of Rad51. (a) Srs2 first translocates in the 3′→5′ direction along an ssDNA overhang to displace Rad51. Srs2 can then repeatedly strip Rad51 from the ssDNA by either maintaining contact with the overhang by creating a small ssDNA loop or remaining at the junction and repeatedly reeling in and releasing the ssDNA (not shown) (93, 110). (b) Rad51 removal from long ssDNA substrates begins with Srs2 loading at small RPA clusters. Rad51 is then stripped from the ssDNA, allowing more RPA to load behind the pioneer Srs2 molecule, which creates more sites for new Srs2 binding events. (c) Srs2 translocates on ssDNA bound by either RPA alone (not shown) or RPA and Rad52 and strips these proteins from the ssDNA. Panels b and c adapted with permission from References 41 and 63. Abbreviations: dsDNA, double-stranded DNA; RPA, replication protein A; ssDNA, single-stranded DNA.
Biophysical Characteristics of Srs2 Antirecombinase Activity
Single-molecule FRET (fluorescence resonance energy transfer) studies have shown Srs2 can undergo repetitive shuttling on short substrates that have an ssDNA/dsDNA junction (110). Similar behaviors have also been reported for Rep, UvrD, PcrA, and other helicases (91–93, 102, 129, 144). An interesting implication of repetitive shuttling is that Srs2 may remain bound to the 3′ ssDNA end while undergoing translocation, resulting in the formation of a small ssDNA loop that enables the enzyme to maintain constant contact with the substrate, although alternative mechanisms may also be possible (Figure 3a) (55, 93). This behavior may enable Srs2 to repeatedly clear Rad51 from ssDNA without dissociating into solution (110).
Single-molecule DNA curtain studies have shown that Srs2 can translocate at a velocity of ~140 nucleotides (nt) s−1, corresponding to the removal of ~50 Rad51 monomers s−1, while traveling for thousands of nucleotides before stopping (63). These studies show that Srs2 loads at RPA clusters present at the ends of Rad51 filaments and it acts in tandem assemblies that travel along the ssDNA in a loosely coordinated manner (Figure 3b) (63). As Srs2 clears Rad51, it creates short tracts of naked ssDNA that are filled in by RPA. Srs2 binds preferentially to RPA–ssDNA rather than Rad51–ssDNA, so the act of removing Rad51 enables more efficient Srs2 loading. Srs2 translocates more rapidly on both naked ssDNA and RPA–ssDNA, exhibiting velocities of ~460 and ~170–190 nt s−1, respectively, compared with the slower velocity (~140 nt s−1) seen on Rad51–ssDNA (41). As a result, Srs2 loaded onto the RPA–ssDNA can catch up to the pioneer helicase at the edge of the receding Rad51 filaments (Figure 3b) (41, 63).
Evidence suggests that Srs2 removes Rad51 by stimulating the ATPase activity of the recombinase (5). Rad51 dissociation (and other Rad51/RecA family members) from ssDNA is coupled to ATP binding and hydrolysis (8, 13, 48, 67, 75, 88, 133). Rad51 requires ATP to bind DNA, whereas ATP hydrolysis and ADP + Pi release allow Rad51 to dissociate from ssDNA, with dissociation taking place most prominently at the filament ends. A mutation in the S. cerevisiae Rad51 ATP binding pocket (K191R) allows ATP binding but inhibits ATP hydrolysis and slows Rad51 dissociation from ssDNA (5, 47, 63, 90, 124). Srs2 can remove Rad51–K191R from ssDNA but does so at a reduced rate compared with wild-type Rad51 (5, 63). This observation led to a model suggesting that Srs2 may stimulate the ATP hydrolysis activity of Rad51, in turn provoking its dissociation from ssDNA (5). One implication of this model is that Srs2 may allosterically stimulate Rad51 ATP hydrolysis activity. Although details remain to be elucidated, one testable prediction arising from this model is that perturbation of protein–protein contacts between Srs2 and Rad51 may slow or prevent Rad51 removal by disrupting allosteric communication between the two proteins.
Srs2 also removes RPA and Rad52 from ssDNA (41), neither of which binds to ATP, suggesting that their displacement from ssDNA may differ from the mechanism of Rad51 disruption (Figure 3c). Indeed, RPA and Rad52 can both bind extremely tightly to ssDNA, with lifetimes exceeding ≥2 h (52, 53), yet neither protein hinders Srs2 translocation relative to Rad51-bound ssDNA (41, 63). Indeed, Srs2 can readily remove RPA and Rad52 from ssDNA, thus increasing the flux between the free and bound states, which may allow for redistribution of both proteins (41, 63). There are two classes of models that may apply to Srs2-mediated removal of RPA and Rad52: either passive or active disruption (61). For passive disruption, Srs2 may encounter RPA or Rad52 and then simply take advantage of transient fluctuations in the protein–ssDNA interfaces to move forward. This type of passive mechanism would be akin to how RNA polymerase II moves through nucleosomes (67). Alternatively, Srs2 may use the energy derived from ATP hydrolysis to actively perturb the protein–nucleic acid interfaces of RPA and Rad52.
Srs2 and Second Strand Capture
Srs2 channels HR intermediates through SDSA during mitosis (Figure 1a). Exactly how Srs2 promotes SDSA is not certain, but current data suggest that the ability of Srs2 to disrupt Rad51 filaments shifts the distribution of early HR intermediates to help ensure that only one end of the DSB can productively engage the homologous target while disfavoring second end capture. This model raises the question of whether the two ends of the DSB are equivalent. For instance, although Rad51 is responsible for the initial stand invasion event, Rad52 may be more important for second strand capture (67). In this regard, the ability of Srs2 to remove Rad52 from ssDNA may prevent second strand capture.
Srs2 Regulation
Several HR accessory factors including Rad52, Rad55/57, and the SHU complex may act as negative regulators of Srs2 (12, 25, 76). Cell biological assays have suggested that Rad52 is a negative regulator of Srs2, revealing that localization of Rad52 and Srs2 in DNA repair foci is anticor-related, but its mechanism of action remains unknown (25). Srs2 readily removes Rad52 from RPA–ssDNA (41), but Srs2 has not yet been tested with Rad52-bound Rad51–ssDNA or in combination with other HR factors. Genetic and biochemical experiments have also implicated the Rad51 paralog complex Rad55/57 as a potential Srs2 regulator, but the mechanism of action remains unknown (46, 76). Similarly, the SHU complex, which comprises the Rad51 paralogs Psy3 and Csm2 and the SWIM-domain proteins Shu1 and Shu2, regulates Srs2 through an unknown mechanism (12, 54, 81).
In principle, these negative regulators might function indirectly by altering the assembly characteristics of Rad51 filaments, for instance, by promoting more rapid reassembly of Rad51 filaments to mitigate the effects of Srs2-mediated filament disruption. Such a model would fit with the known characteristics of Rad52 as a recombination mediator that stimulates Rad51 binding to RPA–ssDNA (60, 101, 127). Alternatively, they may directly alter the behavior of Srs2, for instance, by preventing Srs2 from binding to Rad51 filaments or by blocking Srs2 translocation. This direct inhibition model might apply to Rad55/57 and the SHU complex, which are presumed to enhance the stability of Rad51 filaments (67, 127).
Cell biological experiments have shown that Rad51 filaments are disrupted by Srs2 overexpression during meiosis, whereas Dmc1 filaments remain unaffected (114). This surprising result was extended in biochemical and single-molecule studies, indicating Dmc1 directly inhibits Srs2 translocation and does so by shutting down its ATP hydrolysis activity (37). Furthermore, these data suggest that Srs2 can discriminate between Rad51 and Dmc1 and that interactions with Dmc1 may allosterically regulate the ATPase active site of Srs2 (37). As indicated above, Srs2 promotes SDSA during mitotic growth, favoring the formation of noncrossover recombination products (59), whereas crossovers are favored during meiosis (58). Thus, Dmc1-mediated inhibition of Srs2 may contribute to the shift from noncrossover to crossover recombination during meiosis.
SGS1 IS THE GUARDIAN OF THE GENOME
Sgs1 is a member of the RecQ subfamily of the helicase superfamily 2. The importance of Sgs1 and related RecQ helicases is reflected in the severity of the diseases associated with human RecQ mutations (11, 19, 35, 71). For instance, mutations in BLM (Bloom syndrome helicase), a human homolog of Sgs1, cause Bloom syndrome, which is characterized by extreme UV sensitivity, extensive chromosomal rearrangements, and a predisposition to cancers (6, 15, 44, 49–51). Bloom syndrome patients are so susceptible to DNA damage that they cannot be subjected to normal chemotherapeutic or radiation treatments without risk of secondary cancers (28). Given that yeast Sgs1 and human BLM are closely related, Sgs1 has provided an important model system for understanding how RecQ helicases function in humans.
Biochemical Characteristics of Sgs1
Sgs1 is a large protein (1,447 amino acids, ~166 kDa) that interacts with the proteins Top3 (topoi-somerase III) and Rmi1 (RecQ-mediated genome instability), forming the STR (Sgs1–Top3–Rmi1) complex (11); these accessory factors likely participate in HJ dissolution (see below). Initial biochemical studies of Sgs1 focused on a truncated protein encompassing amino acid residues 400–1,268 (9, 10). This truncated construct contained the core helicase region but lacked the N-terminal domain necessary for protein–protein interactions with Top3 and the C-terminal region that is responsible for Rad51 interactions (9–11). The Sgs1 helicase core exhibited DNA-dependent ATP hydrolysis activity (kcat ~ 10 s−1) and bound preferentially to either plasmid-length ssDNA or forked DNA structures. The Sgs1 core domain has 3′→5′ helicase activity, but it could efficiently unwind only DNA fragments ≤140 base pairs (bp) in length (9, 10). More recently, biochemical studies of full-length Sgs1 revealed that it has more robust DNA-dependent ATP hydrolysis activity [kcat ~ 260 s−1 with poly(dT)], binds tightly to a number of different DNA structures (Kd ~ 0.1–3.0 nM), translocates on DNA in the 3′→5′ direction, and can unwind long dsDNA substrates (up to ~23 kbp) (31).
Sgs1 and DNA End Resection
Sgs1 participates in one of the two primary pathways for DNA end resection (Figure 4a) (reviewed in 40, 67, 125, 126). Sgs1 works with the Top3–Rmi1 complex, the MRX complex (Mre11–Rad50–Xrs2), and Dna2 (30, 97). MRX binds DSB ends and performs initial nucleolytic processing steps to create a loading platform for Sgs1 and Dna2 (40, 67, 125, 126). Dna2 contains a 5′→3′ helicase activity and 3′ and 5′ exo/endonuclease activities (40, 67, 125, 126). The nuclease activities of Dna2 are essential for end resection, but its helicase activity is not (40, 67, 125, 126). Dna2 is regulated by RPA, which suppresses the 3′→5′ nuclease activity, allowing for 5′→3′ resection of the DNA ends (40, 67, 125, 126). Thus, current models for end resection suggest that Sgs1 provides the helicase activity, whereas Dna2 cleaves the DNA (40, 67, 125, 126). As resection proceeds, RPA is loaded onto the newly generated ssDNA, yielding an RPA–ssDNA complex that provides the initial binding substrate for Rad51. Sgs1 interacts with Rad51, but the significance of this interaction remains unknown (11). One possibility is that Sgs1 helps load Rad51 during DNA end resection, similar to what takes place with the bacterial end resection machineries (67). Alternatively, interaction with Rad51 targets Sgs1 to other HR intermediates.
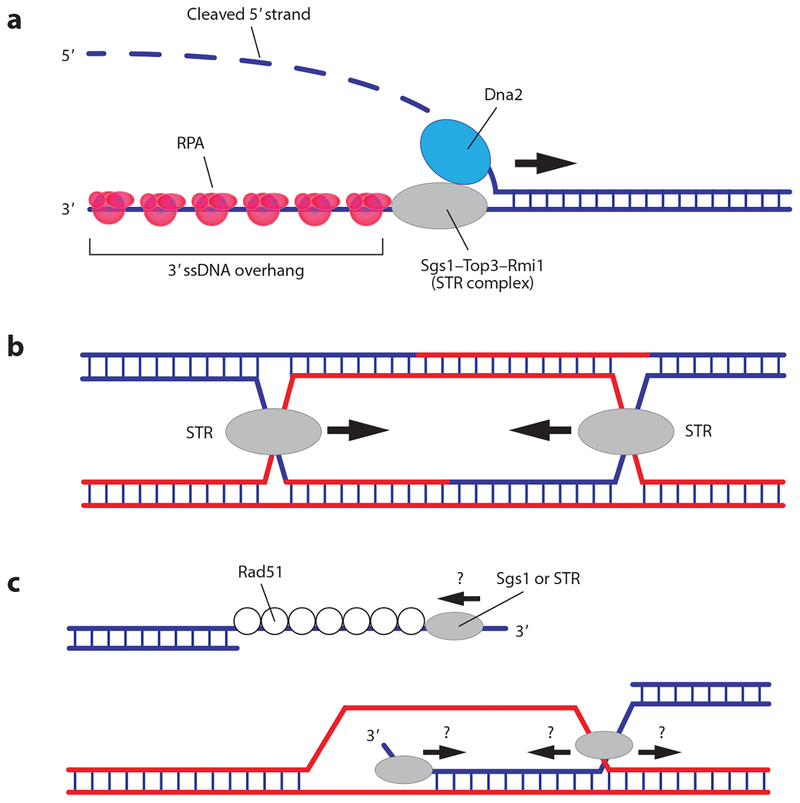
Figure 4.
Sgs1 is the motor that acts during the first and last steps of HR. (a) Sgs1 participates in DNA end resection, functioning as the motor that drives strand separation and propels the resection machinery along the double-stranded DNA. (b) The STR complex is also necessary for later stages of HR, and it may serve as the motor complex that drives convergent branch migration. (c) Although less well understood, Sgs1 (and related RecQ helicases) may also interact with other intermediates, such as the Rad51 presynaptic complex and/or D-loops. Abbreviations: HR, homologous recombination; RPA, replication protein A; ssDNA, single-stranded DNA; STR, Sgs1–Top3–Rmi1.
Sgs1-dependent end resection has been biochemically reconstituted (30, 97), but there remain many open questions. For instance, how is Sgs1 recruited to DNA ends, and how are the activities of Sgs1 and Dna2 coordinated? Is Top3–Rmi1 simply a structural component of the STR complex during end resection, or is it playing a more active role to regulate Sgs1 activity? Does the STR complex assist in loading RPA, Rad51, or other HR proteins onto the newly generated ssDNA overhangs, or are these proteins targeted to the ssDNA through other means? Chromatin and chromatin remodelers clearly influence resection, but details remain to be fully defined (40, 125). Interestingly, although end resection can span up to ~50 kb in vivo, this type of long-range resection occurs only when the break cannot be repaired, so questions remain regarding how resection length is regulated (125, 147). Future studies may help to address these and other questions.
Sgs1–Top3–Rmi1 and Holliday Junction Dissolution
The dHJ is an intermediate in the classical HR pathway (Figure 1a), and this four-stranded DNA structure must be untangled (decatenated) before the completion of HR. Structure-specific nucleases termed HJ resolvases can cleave dHJs to yield crossover or noncrossover products (Figure 1a) (reviewed in 136, 141). Alternatively, dHJ dissolution by the STR complex yields noncrossover products (Figure 1a) (14, 67). During mitosis, dHJ dissolution is preferred because crossover products can cause gross chromosomal rearrangements (14, 67). Consequently, sgs1Δ, top3Δ, and rmi1Δ mutants are all characterized by high crossover frequencies and a propensity for gross chromosomal rearrangements (14, 67, 73). Although the mechanism of HJ dissolution remains unclear, it may involve convergent migration of both HJs, followed by Top3-dependent decatenation of the DNA strands (Figure 4b). Convergent branch migration could take place through thermally driven HJ diffusion or through the ATP-driven translocation activity of Sgs1 (14). Accordingly, Sgs1 and related RecQ helicases can drive HJ branch migration in vitro (22, 31, 36, 64); thus Sgs1 may provide a motor activity to promote convergent branch migration (14). Biochemical studies have suggested that Top3–Rmi1 requires a region of ssDNA to catalyze DNA decatenation and that Sgs1 helicase activity plays an important role in generating this ssDNA substrate (14). Thus, Sgs1 is a critical DNA helicase that functions during all stages of HR, and it (and/or related RecQ helicases) may also act upon other HR intermediates (Figure 4c).
MPH1 AND D-LOOP DISRUPTION
Mph1 is the yeast homolog of human FANCM (Fanconi anemia complementation group M) (137, 142). Highlighting the importance of these proteins, FANCM patients exhibit severe congenital defects, acute myeloid leukemia, and bone marrow failure (16). Mph1 is perhaps the least well understood of the HR-related helicases. Biochemical studies have shown that Mph1 (993 amino acids, 114 kDa) is a 3′→5′ superfamily 2 DeXDD box helicase that exhibits DNA-dependent ATPase activity (kcat ~ 25 s−1) (105, 115). Mph1 can unwind a variety of DNA structures in vitro, including structures containing 3′ or 5′ flaps, branched structures mimicking replication forks, and D-loops (62, 105, 146). Notably, Mph1 and Srs2 can disrupt D-loops in vitro; however, only Mph1 can disrupt D-loops prepared in the presence of Rad54, suggesting that its primary role may be to dismantle D-loops (106). Consistent with its role in HR and similar to what is observed with srs2Δ mutants, mph1Δ mutants exhibit hyper-recombination phenotypes and an elevated frequency of crossovers during mitotic growth (7, 103, 106). Moreover, mph1Δ srs2Δ double mutants have an additive effect on mitotic crossover frequency, suggesting these two proteins act through different mechanisms. These data support a model in which Srs2 and Mph1 both promote SDSA during mitotic growth, with the former acting prior to and the latter acting after strand invasion.
RAD54 AND RDH54 ACT AT MULTIPLE STEPS DURING HOMOLOGOUS RECOMBINATION
The presynaptic complex must perform the homology search and strand invasion reactions (Figure 1) (reviewed in 29, 56, 85, 128). Many questions regarding homology search and strand invasion remain to be addressed, in particular with respect to the contributions of HR accessory factors such as Rad54 and Rdh54.
Biochemical and Biophysical Properties of Rad54 and Rdh54
Rad54 and Rdh54 (also called Tid1) are closely related Swi/Snf-like proteins that belong to the superfamily 2 helicase family (29, 56, 85, 128). Rad54 and Rdh54 core domains are closely related (38% identity and 65% similarity), although their N-terminal domains (~250 amino acids) are unrelated (29). Both proteins are dsDNA-dependent ATPases (Rad54 kcat ~ 20 s−1, Rdh54 kcat ~ 25 s−1), but neither can separate dsDNA strands (84, 95). Instead, Rad54 and Rdh54 translocate along dsDNA at velocities of ~300 and ~80 bp s−1, respectively (4, 95, 108). Rad54 and Rdh54 also stabilize the presynaptic complex, destabilize dsDNA-bound Rad51, promote strand invasion, catalyze branch migration, remodel nucleosomes, and promote strand invasion on chromatin substrates (2, 3, 23, 24, 34, 57, 70, 83, 96, 111, 117, 140, 145). Indeed, it has been difficult to determine precisely what these proteins are doing during HR because of their range of activities.
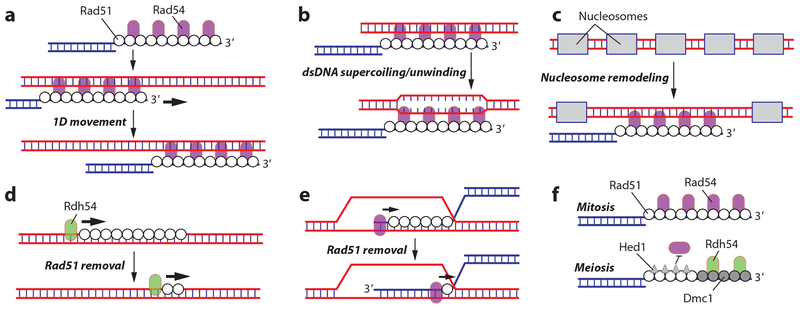
The motor activity of Rad54 may promote the homology search, albeit through poorly understood mechanisms (29, 56, 85, 128). For brevity, we assume that Rdh54 functions similarly to Rad54, consistent with their partial genetic redundancy (29, 56, 85, 128), although we acknowledge that this simplified depiction of Rdh54 may belie its true significance. Several models have been proposed to explain how Rad54 may promote the homology search (reviewed in 29). Rad54 may propel the presynaptic complex along a target dsDNA, thus allowing for a 1D search mechanism (Figure 5a). Rad54 may stimulate dsDNA unwinding, perhaps by altering dsDNA superhelicity, which would promote pairing interactions between the Rad51-bound ssDNA and potential dsDNA targets (Figure 5b). Rad54 also exhibits nucleosome remodeling activity, which may be necessary for the presynaptic complex to interact with chromatin (Figure 5c). All three models are attractive and may contribute to HR, but they remain unproven (29).
Figure 5.
Contributions of Rad54 to homologous recombination. Rad54 may participate in the homology search by (a) propelling the presynaptic complex along a target dsDNA, (b) stimulating dsDNA unwinding, or (c) remodeling or removing nucleosomes bound to dsDNA. (d) Rad54 or its homolog Rdh54 is required for removal of aberrant Rad51 filaments from dsDNA. (e) Removal of Rad51 from the heteroduplex recombination product by Rad54 allows the DNA replication machinery to access the 3′OH of the invading strand. (f) Differential regulation of Rad51 and Dmc1 through Hed1-mediated inhibition of Rad54, possibly allowing for Rdh54 to stimulate Dmc1-specific strand invasion. Abbreviation: dsDNA, double-stranded DNA.
Rad54 and Rdh54 Disrupt Rad51–dsDNA Filaments
Rad51 and Dmc1 can bind tightly to dsDNA, which has the potential to create a pathological situation. Importantly, Rad54 and Rdh54 can strip Rad51 and Dmc1 from dsDNA through use of their ATP-dependent dsDNA translocation activities (57, 82, 117). Rdh54 removes Dmc1 from undamaged chromatin in meiotic cells, and Rad54 can substitute for this activity when Rdh54 is absent (Figure 5d) (57). In the absence of Rdh54, Dmc1 accumulates on chromatin and appears as bright fluorescent foci in microscopic analysis of chromosome spreads (57). Removal of these foci requires the ATP hydrolysis activity of Rdh54, suggesting that it depends on Rdh54 motor activity. Rad51 overexpression during mitosis causes accumulation of nonproductive Rad51 on chromatin, leading to chromosome instability (117). RAD51 is commonly overexpressed in human cancers, so the toxicity of Rad51 overexpression in yeast may mimic some aspects of human pathology. Rad54 and another closely related protein called Uls1 also contribute to Rad51 removal, albeit to a lesser extent than Rdh54 (117). Although these studies show that Rad54 and Rdh54 prevent accumulation of Rad51 and Dmc1 on undamaged dsDNA, this activity has not yet been carefully studied in vitro.
Rad51 and Dmc1 must also be removed from the heteroduplex dsDNA after strand invasion to allow for subsequent steps in the repair reaction. Biochemical studies have provided a satisfying model for understanding how Rad54 removes Rad51 from the heteroduplex dsDNA product of strand invasion (140). In this model, Rad54 initially interacts with the presynaptic complex through the Rad54 N-terminal domain, which binds to ssDNA (140). Following strand invasion, the Rad54 motor domain binds the resulting dsDNA and then translocates along the heteroduplex dsDNA while removing Rad51 (Figure 5e) (140). Given their similarities, Rdh54 may function in a similar manner with Dmc1.
Rad54 Is a Key Regulatory Control Point in Meiosis
Rad54 and Rdh54 have overlapping functions. However, Rad54 may be more important for intersister HR during mitotic growth in haploids, whereas Rdh54 may be more important for interhomolog HR during mitotic growth in diploids and during meiosis. Regulatory features that control Rad54 and Rdh54 specificities for interhomolog versus intersister HR are best understood within meiosis. Rad54 is a required cofactor for Rad51 activity in vivo. However, during meiosis, Rad54–Rad51 interactions are weakened by Mek1-dependent Rad54 phosphorylation (99) and by Hed1, which binds tightly to Rad51 and prevents association of Rad54 (Figure 5f) (26, 27, 38, 130). These regulatory interactions do not affect Rad54 interactions with Dmc1, nor do they appear to affect Rdh54. Biochemical data also suggest that Rdh54 interacts preferentially with Dmc1 (96). Combining these effects ensures that Dmc1 is the catalytically active recombinase for interhomolog recombination during meiosis. There is as yet no explanation for why Rad54 and Rdh54 differentially affect intersister and interhomolog recombination during mitosis.
CONCLUSION
We have briefly highlighted the S. cerevisiae helicases that participate in HR. The extensive literature on the genetic consequences of their perturbation provides an advantage to studying yeast helicases, thus offering a crucial framework for guiding both biochemical and biophysical studies. Moreover, each of these proteins has one or more putative human homologs, many of which are associated with severe genetic disorders, and mutations in the yeast proteins often phenocopy these disorders. Thus, studies of the yeast proteins can yield direct insights into the molecular defects underlying human disease. As illustrated throughout our discussion, we now know much about basic helicase functions, but fully understanding how they are integrated into the HR pathway remains challenging. In moving forward, key questions will focus on the mechanistic details by which helicases interact with HR intermediates and how these interactions are influenced by other accessory proteins.
ACKNOWLEDGMENTS
Research in the Greene laboratory is funding by NIH grants R35GM118026 and P01CA92584 (E.C.G.), a Wellcome Trust Collaborative Grant (E.C.G.), and NSF grant MCB-1817315 (E.C.G.). J.B.C. is the Mark Foundation for Cancer Research Fellow for the Damon Runyon Cancer Research Foundation (DRG 2310–17).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aguilera A, Klein HL. 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119:779–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeev A, Mazin A, Kowalczykowski SC. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Mol. Biol 10:182–86 [DOI] [PubMed] [Google Scholar]
- 3.Alexiadis V, Kadonaga JT. 2002. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16:2767–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amitani I, Baskin RJ, Kowalczykowski SC. 2006. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell 23:143–48 [DOI] [PubMed] [Google Scholar]
- 5.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. 2009. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 35:105–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arora H, Chacon AH, Choudhary S, McLeod MP, Meshkov L, et al. 2014. Bloom syndrome. Int. J. Dermatol 53:798–802 [DOI] [PubMed] [Google Scholar]
- 7.Banerjee S, Smith S, Oum JH, Liaw HJ, Hwang JY, et al. 2008. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J. Cell Biol 181:1083–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. 2012. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491:274–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett RJ, Keck JL, Wang JC. 1999. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol 289:235–48 [DOI] [PubMed] [Google Scholar]
- 10.Bennett RJ, Sharp JA, Wang JC. 1998. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem 273:9644–50 [DOI] [PubMed] [Google Scholar]
- 11.Bernstein KA, Gangloff S, Rothstein R. 2010. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet 44:393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. 2011. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 antirecombinase. Mol. Biol. Cell 22:1599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianco PR, Tracy RB, Kowalczykowski SC. 1998. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci 3:D570–603 [DOI] [PubMed] [Google Scholar]
- 14.Bizard AH, Hickson ID. 2014. The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol 6:a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom D 1954. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs. Probably a syndrome entity. AMA Am. J. Dis. Child 88:754–58 [PubMed] [Google Scholar]
- 16.Bogliolo M, Surralles J. 2015. Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr. Opin. Genet. Dev 33:32–40 [DOI] [PubMed] [Google Scholar]
- 17.Branzei D, Foiani M. 2007. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 21:3019–26 [DOI] [PubMed] [Google Scholar]
- 18.Branzei D, Szakal B. 2017. Building up and breaking down: mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol 52:381–94 [DOI] [PubMed] [Google Scholar]
- 19.Brosh RM Jr. 2013. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13:542–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosh RM Jr., Bohr VA. 2007. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 35:7527–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown MS, Bishop DK. 2014. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol 7:a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bugreev DV, Brosh RM Jr., Mazin AV. 2008. RECQ1 possesses DNA branch migration activity. J. Biol. Chem 283:20231–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugreev DV, Hanaoka F, Mazin AV. 2007. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol 14:746–53 [DOI] [PubMed] [Google Scholar]
- 24.Bugreev DV, Mazina OM, Mazin AV. 2006. Rad54 protein promotes branch migration of Holliday junctions. Nature 442:590–93 [DOI] [PubMed] [Google Scholar]
- 25.Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R. 2009. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol 185:969–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busygina V, Saro D, Williams G, Leung WK, Say AF, et al. 2012. Novel attributes of Hed1 affect dynamics and activity of the Rad51 presynaptic filament during meiotic recombination. J. Biol. Chem 287:1566–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22:786–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell MB, Campbell WC, Rogers J, Rogers N, Rogers Z, et al. 2018. Bloom syndrome: research and data priorities for the development of precision medicine as identified by some affected families. Cold Spring Harb. Mol. Case Stud 4:a002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceballos SJ, Heyer WD. 2011. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta 1809:509–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, et al. 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467:112–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cejka P, Kowalczykowski SC. 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J. Biol. Chem 285:8290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Wold MS. 2014. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays 36:1156–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chester N, Kuo F, Kozak C, O’Hara CD, Leder P. 1998. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom’s syndrome gene. Genes Dev. 12:3382–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi P, Kwon Y, Seong C, Epshtein A, Lam I, et al. 2006. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J. Biol. Chem 281:26268–79 [DOI] [PubMed] [Google Scholar]
- 35.Chu WK, Hickson ID. 2009. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer 9:644–54 [DOI] [PubMed] [Google Scholar]
- 36.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, et al. 2000. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crickard JB, Kaniecki K, Kwon Y, Sung P, Greene EC. 2018. Meiosis-specific recombinase Dmc1 is a potent inhibitor of the Srs2 antirecombinase. PNAS 115:E10041–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crickard JB, Kaniecki K, Kwon Y, Sung P, Lisby M, Greene EC. 2018. Regulation of Hed1 and Rad54 binding during maturation of the meiosis-specific presynaptic complex. EMBO J. 37:e98728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croteau DL, Popuri V, Opresko PL, Bohr VA. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem 83:519–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daley JM, Niu H, Miller AS, Sung P. 2015. Biochemical mechanism of DSB end resection and its regulation. DNA Repair 32:66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Tullio L, Kaniecki K, Kwon Y, Crickard JB, Sung P, Greene EC. 2017. Yeast Srs2 helicase promotes redistribution of single-stranded DNA-bound RPA and Rad52 in homologous recombination regulation. Cell Rep. 21:570–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deans B, Griffin CS, Maconochie M, Thacker J. 2000. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 19:6675–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elango R, Sheng Z, Jackson J, DeCata J, Ibrahim Y, et al. 2017. Break-induced replication promotes formation of lethal joint molecules dissolved by Srs2. Nat. Commun 8:1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, et al. 1995. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83:655–66 [DOI] [PubMed] [Google Scholar]
- 45.Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, et al. 2011. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. PNAS 108:686–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fortin GS, Symington LS. 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 21:3160–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fung CW, Fortin GS, Peterson SE, Symington LS. 2006. The rad51-K191R ATPase-defective mutant is impaired for presynaptic filament formation. Mol. Cell. Biol 26:9544–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. 2006. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443:875–78 [DOI] [PubMed] [Google Scholar]
- 49.German J 1997. Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet. Cytogenet 93:100–6 [DOI] [PubMed] [Google Scholar]
- 50.German J, Archibald R, Bloom D. 1965. Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science 148:506–7 [DOI] [PubMed] [Google Scholar]
- 51.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. 2007. Syndrome-causing mutations of the BLM gene in persons in the Bloom’s Syndrome Registry. Hum. Mutat 28:743–53 [DOI] [PubMed] [Google Scholar]
- 52.Gibb B, Ye LF, Gergoudis SC, Kwon Y, Niu H, et al. 2014. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLOS ONE 9:e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibb B, Ye LF, Kwon Y, Niu H, Sung P, Greene EC. 2014. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol 21:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godin S, Wier A, Kabbinavar F, Bratton-Palmer DS, Ghodke H, et al. 2013. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 41:4525–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ha T, Kozlov AG, Lohman TM. 2012. Single-molecule views of protein movement on single-stranded DNA. Annu. Rev. Biophys 41:295–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heyer WD, Li X, Rolfsmeier M, Zhang XP. 2006. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 34:4115–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holzen TM, Shah PP, Olivares HA, Bishop DK. 2006. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 20:2593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunter N 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jasin M, Rothstein R. 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. 2007. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129:1299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang YH, Munashingha PR, Lee CH, Nguyen TA, Seo YS. 2012. Biochemical studies of the Saccharomyces cerevisiae Mph1 helicase on junction-containing DNA structures. Nucleic Acids Res. 40:2089–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaniecki K, De Tullio L, Gibb B, Kwon Y, Sung P, Greene EC. 2017. Dissociation of Rad51 presynaptic complexes and heteroduplex DNA joints by tandem assemblies of Srs2. Cell Rep. 21:3166–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. 2000. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. PNAS 97:6504–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kass EM, Moynahan ME, Jasin M. 2016. When genome maintenance goes badly awry. Mol. Cell 62:777–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keeney S, Lange J, Mohibullah N. 2014. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet 48:187–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kowalczykowski SC. 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol 7:016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, et al. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305–9 [DOI] [PubMed] [Google Scholar]
- 69.Kuznetsov SG, Haines DC, Martin BK, Sharan SK. 2009. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 69:863–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon Y, Seong C, Chi P, Greene EC, Klein H, Sung P. 2008. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J. Biol. Chem 283:10445–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsen NB, Hickson ID. 2013. RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol 767:161–84 [DOI] [PubMed] [Google Scholar]
- 72.Lee JY, Yang W. 2006. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127:1349–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, et al. 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19:339–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim DS, Hasty P. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol 16:7133–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindsley JE, Cox MM. 1989. Dissociation pathway for recA nucleoprotein filaments formed on linear duplex DNA. J. Mol. Biol 205:695–711 [DOI] [PubMed] [Google Scholar]
- 76.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. 2011. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479:245–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lohman TM, Tomko EJ, Wu CG. 2008. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol 9:391–401 [DOI] [PubMed] [Google Scholar]
- 78.Lok BH, Carley AC, Tchang B, Powell SN. 2013. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32:3552–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lorenz A 2017. Modulation of meiotic homologous recombination by DNA helicases. Yeast 34:195–203 [DOI] [PubMed] [Google Scholar]
- 80.Marini V, Krejci L. 2010. Srs2: the “Odd-Job Man” in DNA repair. DNA Repair 9:268–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martino J, Bernstein KA. 2016. The Shu complex is a conserved regulator of homologous recombination. FEMS Yeast Res. 16:fow073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mason JM, Dusad K, Wright WD, Grubb J, Budke B, et al. 2015. RAD54 family translocases counter genotoxic effects of RAD51 in human tumor cells. Nucleic Acids Res. 43:3180–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mazin AV, Alexeev AA, Kowalczykowski SC. 2003. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem 278:14029–36 [DOI] [PubMed] [Google Scholar]
- 84.Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. 2000. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 6:583–92 [DOI] [PubMed] [Google Scholar]
- 85.Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. 2010. Rad54, the motor of homologous recombination. DNA Repair 9:286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCarthy EE, Celebi JT, Baer R, Ludwig T. 2003. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol. Cell. Biol 23:5056–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mehta A, Haber JE. 2014. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Menetski JP, Kowalczykowski SC. 1985. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol 181:281–95 [DOI] [PubMed] [Google Scholar]
- 89.Morrical SW. 2015. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol 7:a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morrison C, Shinohara A, Sonoda E, Yamaguchi-Iwai Y, Takata M, et al. 1999. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol 19:6891–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myong S, Bruno MM, Pyle AM, Ha T. 2007. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317:513–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, et al. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323:1070–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. 2005. Repetitive shuttling of a motor protein on DNA. Nature 437:1321–25 [DOI] [PubMed] [Google Scholar]
- 94.Neale MJ, Keeney S. 2006. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442:153–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. 2007. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J. Biol. Chem 282:30776–84 [DOI] [PubMed] [Google Scholar]
- 96.Nimonkar AV, Dombrowski CC, Siino JS, Stasiak AZ, Stasiak A, Kowalczykowski SC. 2012. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J. Biol. Chem 287:28727–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467:108–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niu H, Klein HL. 2017. Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 17:fow111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Niu H, Wan L, Busygina V, Kwon Y, Allen JA, et al. 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palladino F, Klein HL. 1992. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132:23–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paques F, Haber JE. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev 63:349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, et al. 2010. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell 142:544–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Piazza A, Wright WD, Heyer WD. 2017. Multi-invasions are recombination byproducts that induce chromosomal rearrangements. Cell 170:760–73.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pittman DL, Schimenti JC. 2000. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis 26:167–73 [DOI] [PubMed] [Google Scholar]
- 105.Prakash R, Krejci L, Van Komen S, Anke Schurer K, Kramer W, Sung P. 2005. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem 280:7854–60 [DOI] [PubMed] [Google Scholar]
- 106.Prakash R, Satory D, Dray E, Papusha A, Scheller J, et al. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. 2007. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J. Mol. Biol 369:940–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pyle AM. 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys 37:317–36 [DOI] [PubMed] [Google Scholar]
- 110.Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. 2013. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun 4:2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ristic D, Wyman C, Paulusma C, Kanaar R. 2001. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. PNAS 98:8454–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rong L, Klein HL. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem 268:1252–59 [PubMed] [Google Scholar]
- 113.Rong L, Palladino F, Aguilera A, Klein HL. 1991. The hyper-gene conversion hpr5–1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127:75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sasanuma H, Furihata Y, Shinohara M, Shinohara A. 2013. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase. Genetics 194:859–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scheller J, Schurer A, Rudolph C, Hettwer S, Kramer W. 2000. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155:1069–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Seki M, Otsuki M, Ishii Y, Tada S, Enomoto T. 2008. RecQ family helicases in genome stability: lessons from gene disruption studies in DT40 cells. Cell Cycle 7:2472–78 [DOI] [PubMed] [Google Scholar]
- 117.Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL. 2010. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol. Cell 39:862–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, et al. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804–10 [DOI] [PubMed] [Google Scholar]
- 119.Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. 1999. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53−/− background. Mol. Cell. Biol 19:8686–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem 76:23–50 [DOI] [PubMed] [Google Scholar]
- 121.Singleton MR, Wigley DB. 2002. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol 184:1819–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, et al. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Soultanas P, Wigley DB. 2000. DNA helicases: ‘inching forward.’ Curr. Opin. Struct. Biol 10:124–28 [DOI] [PubMed] [Google Scholar]
- 124.Sung P, Stratton SA. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem 271:27983–86 [DOI] [PubMed] [Google Scholar]
- 125.Symington LS. 2014. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol 6:a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet 45:247–71 [DOI] [PubMed] [Google Scholar]
- 127.Symington LS, Rothstein R, Lisby M. 2014. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198:795–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tan TL, Kanaar R, Wyman C. 2003. Rad54, a Jack of all trades in homologous recombination. DNA Repair 2:787–94 [DOI] [PubMed] [Google Scholar]
- 129.Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM. 2010. 5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 29:3826–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsubouchi H, Roeder GS. 2006. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20:1766–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, et al. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. PNAS 93:6236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Van Komen S, Reddy MS, Krejci L, Klein H, Sung P. 2003. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem 278:44331–37 [DOI] [PubMed] [Google Scholar]
- 133.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. 2009. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature 457:745–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309–12 [DOI] [PubMed] [Google Scholar]
- 135.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75–84 [DOI] [PubMed] [Google Scholar]
- 136.West SC, Blanco MG, Chan YW, Matos J, Sarbajna S, Wyatt HD. 2015. Resolution of recombination intermediates: mechanisms and regulation. Cold Spring Harb. Symp. Quant. Biol 80:103–9 [DOI] [PubMed] [Google Scholar]
- 137.Whitby MC. 2010. The FANCM family of DNA helicases/translocases. DNA Repair 9:224–36 [DOI] [PubMed] [Google Scholar]
- 138.White RR, Vijg J. 2016. Do DNA double-strand breaks drive aging? Mol. Cell 63:729–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem 66:61–92 [DOI] [PubMed] [Google Scholar]
- 140.Wright WD, Heyer WD. 2014. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell 53:420–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wyatt HD, West SC. 2014. Holliday junction resolvases. Cold Spring Harb. Perspect. Biol 6:a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xue X, Sung P, Zhao X. 2015. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 29:1777–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang W 2010. Lessons learned from UvrD helicase: mechanism for directional movement. Annu. Rev. Biophys 39:367–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. 2009. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 28:405–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Z, Fan HY, Goldman JA, Kingston RE. 2007. Homology-driven chromatin remodeling by human RAD54. Nat. Struct. Mol. Biol 14:397–405 [DOI] [PubMed] [Google Scholar]
- 146.Zheng XF, Prakash R, Saro D, Longerich S, Niu H, Sung P. 2011. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair 10:1034–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134:981–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol 7:a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]