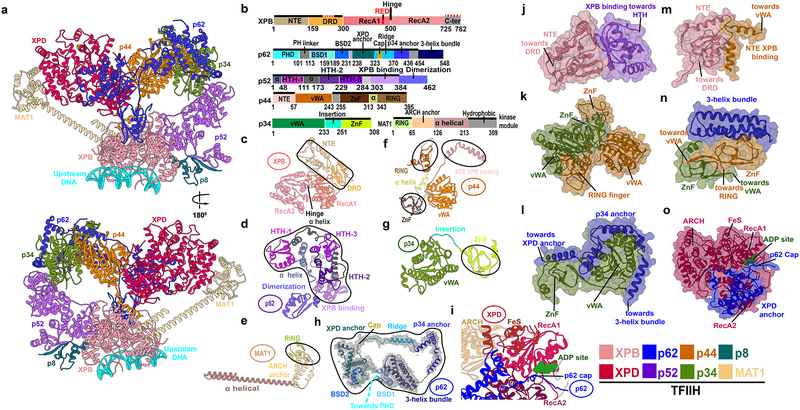
Figure 1. TFIIH integrative structure model based on comparative analysis of cryo-EM densities reveals “molecular rigging” formed by p62 and p44.
a, Anterior and posterior cartoon views of TFIIH GTF where missing regions or entire domains and proteins are built for XPB, p62, p52, p44, p34 and MAT1. Newly modeled p62 and p44 subunits act as molecular rigging, interlinking TFIIH. b, Motif schematic highlighting newly modeled regions (solid black lines). Two small regions in XPB, not present in the EM maps, were not modeled (red dashed lines). Abbreviations denote: DRD - damage recognition domain; NTE - N-terminal extension; HTH- helix-turn-helix. c-h, Cartoon of TFIIH subunits with newly modeled regions circled. h, Representative cryo-EM electron density from apo-TFIIH overlaid with p62 (PHD domain not shown). i, Zoom view of p62 cap region overlaying the XPD ATP binding cleft. Space filling views highlighting Interactions newly modeled in j, XPB NTE and p52; k, p34 and p44; l, p62 helices and p34; m, XPB N-terminus with p44; n, p62 3-helix bundle and p34 plus p44; and o, p62 and XPD.