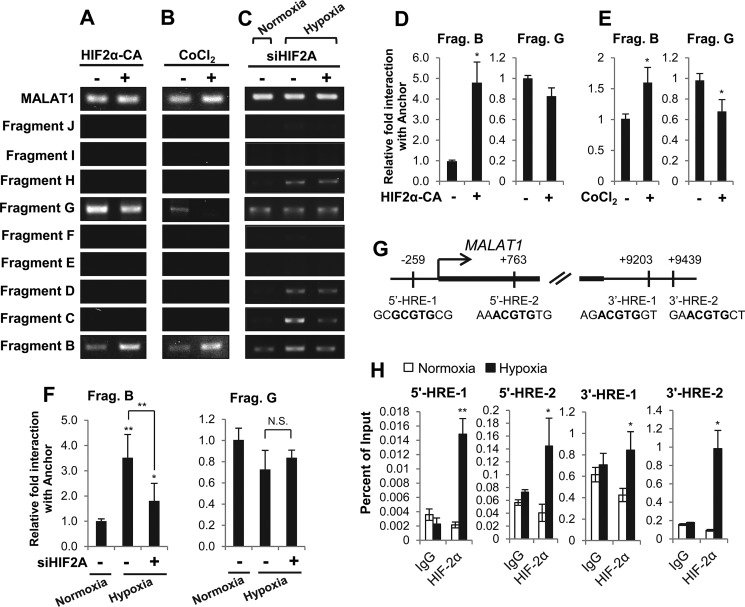
Figure 5.
HIF-2α expression levels modulate chromatin looping between the MALAT1 promoter and the 3′ enhancer in MCF7 breast cancer cells. A–C, gel images of 3C-PCR from MCF7 cells transfected with pCDNA-HIF2α-P405A/P531A (HIF2α-CA; +) or an empty vector (−) for 48 h (A), treated with 100 μm CoCl2 (+) or vehicle (−) for 24 h (B), or transfected with HIF2A siRNA (+) or control siRNA (−) for 48 h prior to an additional 24-h incubation under normoxia or hypoxia as indicated (C). D–F, qPCR analyses of the relative interaction frequencies between the anchor (3′ enhancer) and fragments B or G following 48 h of HIF-2α overexpression (HIF-CA) (D), 24 h of 100 μm CoCl2 treatment (E) or 72 total hours of HIF-2α knockdown as described in C (F). G, diagram of the MALAT1 promoter and the 3′ enhancer with the location and the sequence of HIF-binding motifs (HREs) labeled in bold. Two HREs are predicted within the MALAT1 promoter (5′-HRE-1 and 5′HRE-2) and two are predicted within the putative 3′ enhancer (3′-HRE-1 and 3′ HRE-2). H, ChIP-qPCR demonstrates enrichment of HIF-2α under normoxic or 24-h hypoxic conditions in MCF7 cells. Data with bar graphs represent mean ± S.D., n = 3, *, p < 0.05; **, p < 0.01.