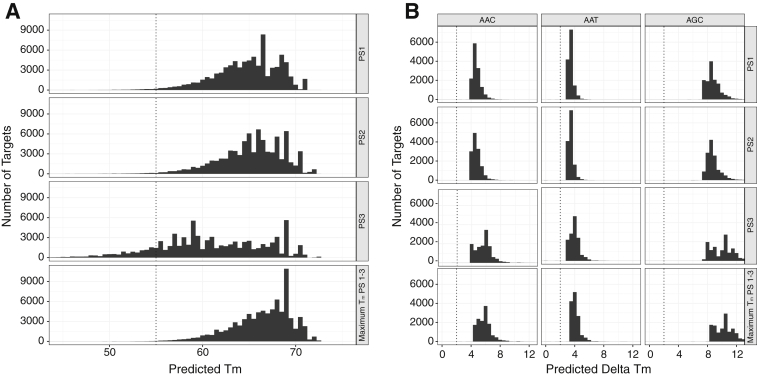
Figure 3.
In silico simulations for probe set selection. A: Distribution of predicted melting temperature (Tm) for 81,126 sequences with three 34-bp probe sets (PS1, PS2, and PS3). Sequences included the 13,521 distinct low- and middle-income country 203-bp database sequences that encompassed codon 103 each with a different one of the six reported codons at position 103 (AAA, AAC, AAG, AAT, AGA, AGC). For each of the 81,126 (13,521 × 6) sequences, max_Tm_PS1-3 indicated the maximum Tm of PS1, PS2, and PS3. The temperature used for initial hybridization was 55°C. Dashed lines are at 55°C, the temperature at which binding is considered necessary for probe capture. B: Distribution of predicted ΔTm between probes complementary to AAC, AAT, and AGC versus probes complementary to AAA for each of the three probe sets (PS1, PS2, and PS3). Because the probes within a probe set shared the same flanking sequence, the ΔTm was caused entirely by the central mismatch at the codon 103 position. Dashed lines are at a ΔTm of 2°C, the minimum temperature considered necessary for distinguishing between the finding of two different targets to the same probe.