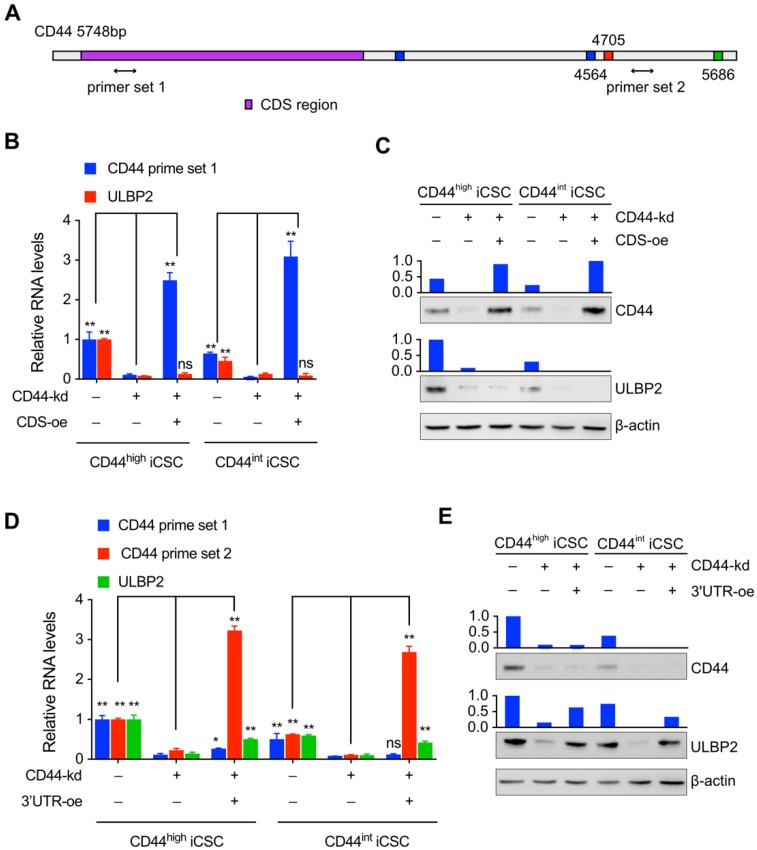
Figure 3.
The regulation of ULBP2 was performed by CD44 3'-untranslated region. (A) Schematic diagram of CD44 genes and the primer sets used in the qPCR assays. (B) CD44highiCSC/CD44intiCSC stably expressing dCas9-KRAB and sgRNA against CD44 (CD44-knockdown, CD44-kd) were transfected with blank expression vector (Ctrl) or CD44 coding sequence expression vector (CD44 CDS-overexpression, CDS-oe). CD44 and ULBP2 transcript levels were determined by qRT-PCR. Levels are represented relative to those found in control-transfected cells as means mean ± SD (n=3) (ns: not significant, **: p < 0.01). (C) CD44 and ULBP2 protein levels of identical cells (Fig. 3B) were analyzed by Western blotting. β-actin served as a loading control. (D) CD44highiCSC/CD44intiCSC stably expressing dCas9-KRAB and sgRNA against CD44 (CD44-knockdown, CD44-kd) were transfected with blank expression vector (Ctrl) or CD44 3'UTR expression vector (CD44 3'UTR -overexpression, 3'UTR-oe). CD44 CDS, CD44 3'UTR, and ULBP2 transcript levels were determined by qRT-PCR. Levels are represented relative to those found in control-transfected cells as means mean ± SD (n=3) (ns: not significant, *:p<0.05, **:p < 0.01). (E) CD44 and ULBP2 protein levels of identical cells (Fig. 3D) were analyzed by Western blotting. β-actin served as a loading control.