Figure 1. Drawing of riboswitch-mediated gene regulation and overview of a riboswitch-mediated GFPuv reporter coupled (ReCo) to in cell (ic)SHAPE.
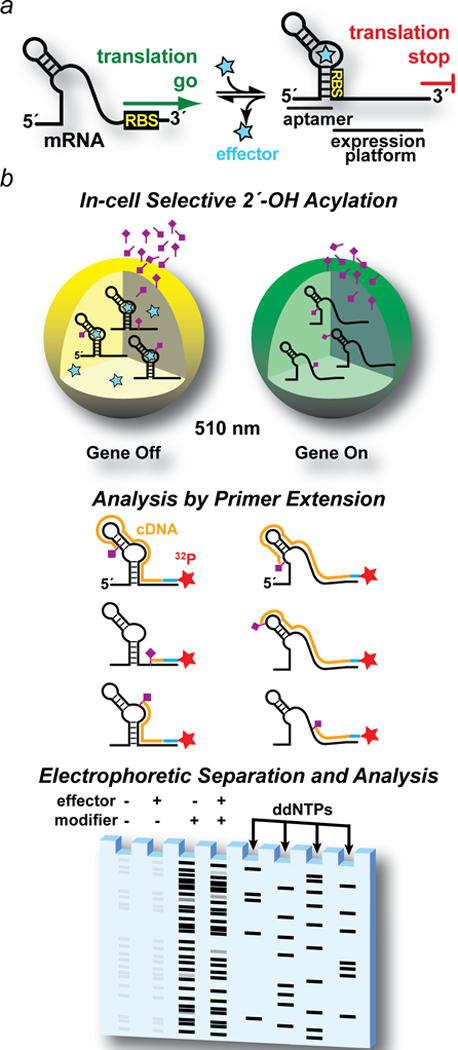
(a) Ideal two-state control of mRNA translation by a riboswitch. When cellular effector levels are low, the ribosome-binding site (RBS) in the 5′-leader is exposed to interact with the ribosome, leading to protein translation (i.e., a gene “on” conformation). When effector levels rise (star), the aptamer domain senses the cognate effector, shifting the conformational equilibrium to a compact state that reduces SDS accessibility in the expression platform, thus attenuating translation (i.e., a gene “off” conformation). (b) Major steps in riboswitch-mediated gene regulation coupled to icSHAPE. Step 1. Cells are grown ± effector to elicit gene “off” and “on” conformational states of the target regulatory RNA. Gene regulation is monitored by GFPuv fluorescence emission at 510 nm recorded using live cells. The RNA modification reagent (tailed rhombus) is incubated with each cell population. Step 2. Cells are lysed to recover RNA. A 32P-labeled primer complementary to the GFPuv gene allows reverse transcription to produce cDNA (orange lines). Primer extension stops at each site of 2´-acylation that occurred in live cells. Step 3. cDNA is separated by size using denaturing polyacrylamide gel electrophoresis. Controls are included for untreated cells (i.e., no chemical modifier ± effector) to correct for factors such as endogenous primer pausing. As a sequence reference, primer extensions are run on unmodified RNA in the presence of ddNTPs to produce a sequence ladder.
