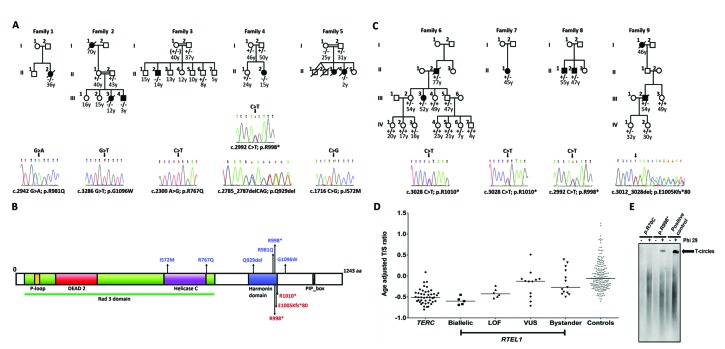
Figure 1.
The segregation, location and impact of RTEL1 variants. (A) Families with biallelic RTEL1 variants and sequencing traces of index cases (homozygous, Families 1, 2, 3 and 5; compound heterozygous, Family 4). The genotyping is described as follows: wild-type (+/+), heterozygous (+/−) or biallelic (−/−). The age at study is given in years. Affected individuals are coloured in black. NA: not available. (B) RTEL1 protein (NP_116575.3) schematic showing the location of the biallelic variants in blue, and the heterozygous LOF variants in red. Conserved protein domains include the P-loop NTPase (yellow); the Rad3 domain (green) which includes the DEAD2 domain (red) and the Helicase C-terminal domain (purple); Harmonin N-like domain (blue); PIP-box – the proliferating cell nuclear antigen interacting protein domain (black). (C) Families with heterozygous LOF RTEL1 variants and sequencing traces of index cases, annotated as in panel A. (D) Age adjusted telomere length values (delta-tel) were measured by subtracting the observed T/S ratio from the expected T/S ratio, using the equation derived from the line of best fit through the plot of T/S ratios from healthy control samples against age. Patients with TERC variants are included as a group with known short telomeres. Centiles were calculated from the control delta-tel values as follows: 99th centile = 0.95, 90th centile = 0.42, 50th centile = 0.06, 10th centile = −0.34, 1st centile = −0.54. The different genotypes are represented as follows, TERC-circles (n=44); biallelic-squares (n=5); loss of function (LOF)-triangles (n=6); variants of unknown significance (VUS)-diamonds (n=12); bystanders-inverted triangles (n=13); controls-grey squares (n=202). (E) T-circle amplification using Phi29 polymerase detected by Southern Blot analysis. Samples: p.R70C - patient with sporadic DC carrying this variant of unknown significance (patient 13 in Table 1); p.R998* - proband of Family 8 carrying this LOF variant (Table 1); positive control - genomic DNA extracted from WI-38 VA-13 cells, known to produce T-circle.