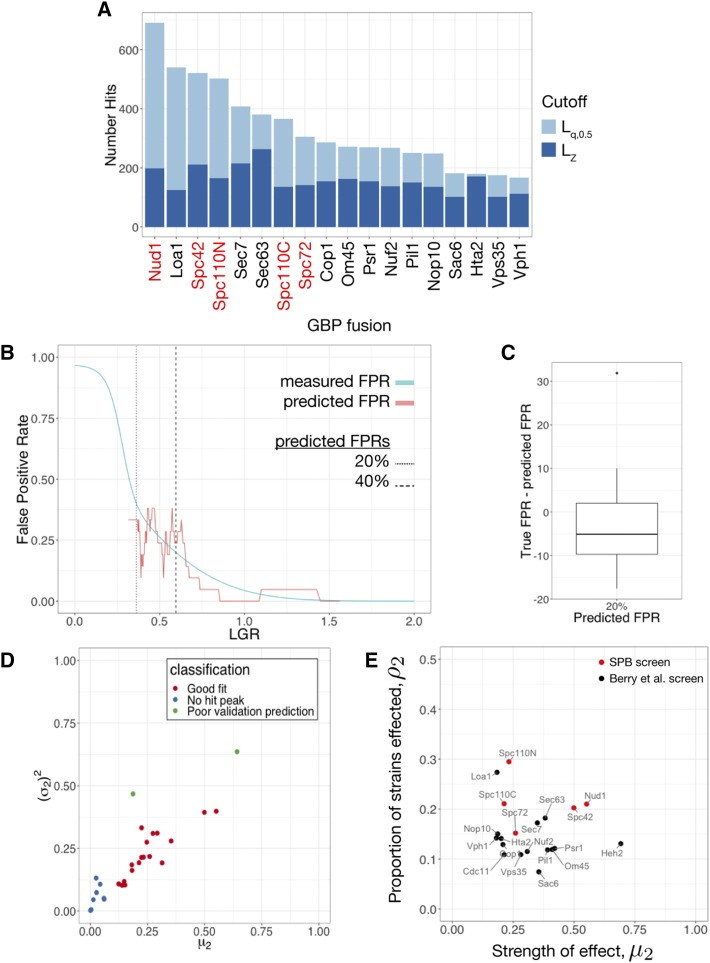
Figure 3.
A: The number of hits by both Z-score () and q(x) () cutoff for each of the screens where the mixture model was applicable. The q(x) cutoff has a higher dynamic range than the Z-score and is better able to distinguish screens with many hits. B: FPR prediction for the Spc72 screen. The FPR for the screen was predicted from the mixture model and this prediction is overlaid with estimates of the FPR using binned data from the validation screen. In this case, the predicted FPR was reasonably accurate, although the data are quite noisy. The points where the mixture model predicts and FPR are indicated with a dashed line. C: Box-and-whisker plot showing the difference between measured and predicted FPR at the point where the FPR is predicted to be across the screens where the mixture model was applicable. This shows some bias, with the predicted FPR generally higher than the true FPR but generally achieving an accuracy around . D: Classification of mixture model fit for each of the 28 screens analyzed. The mean and variance of component 2 are good indicators of the success of the model with very low means or high variances indicative of the lack of a hit peak or poor validation prediction respectively. E: Classification of screen based on fitted parameters calculated using the subset of GFP strains used in the SPB screen. Each of the screens for which the mixture model fit was appropriate are plotted according to the proportion of strains affected () and the average strength of these effects (). The SPB screens Spc42 and Nud1 are positioned in the upper right portion of the graph, showing that a large proportion of proteins were sensitive to forced interaction with the SPB and these sensitivities caused significant growth defects.