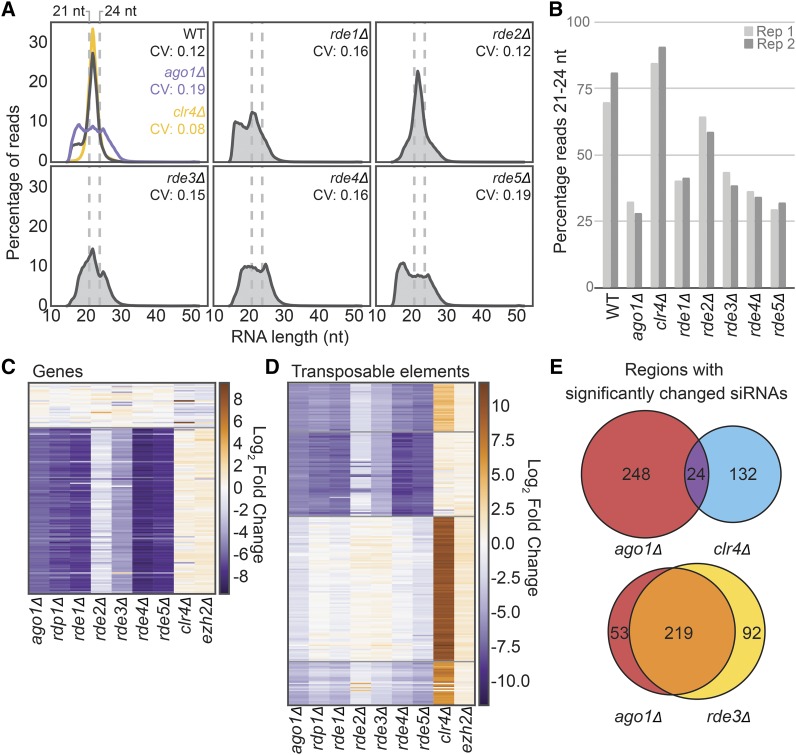
Figure 3.
A. Distribution of small RNA-seq read sizes and coefficient of variation (CV) in each RDE knockout as well as ago1Δ and a partial deletion of clr4Δ (single replicate shown). B. Percentage of small RNA-seq reads between 21-24 nt in length for each knockout compared to wild type. C. Fold change of siRNA abundance for annotated genes with at least 100 small RNA-seq reads in both of the wild-type libraries (determined by DESeq2, 2 replicates). Order determined by K-means clustering. D. Fold change of siRNA abundance (determined by DESeq2, 2 replicates) against transposable elements and transposable element remnants (see methods for annotation strategy) compared to wild type. E. Overlap of transposable elements and genes with significantly changed siRNA populations.