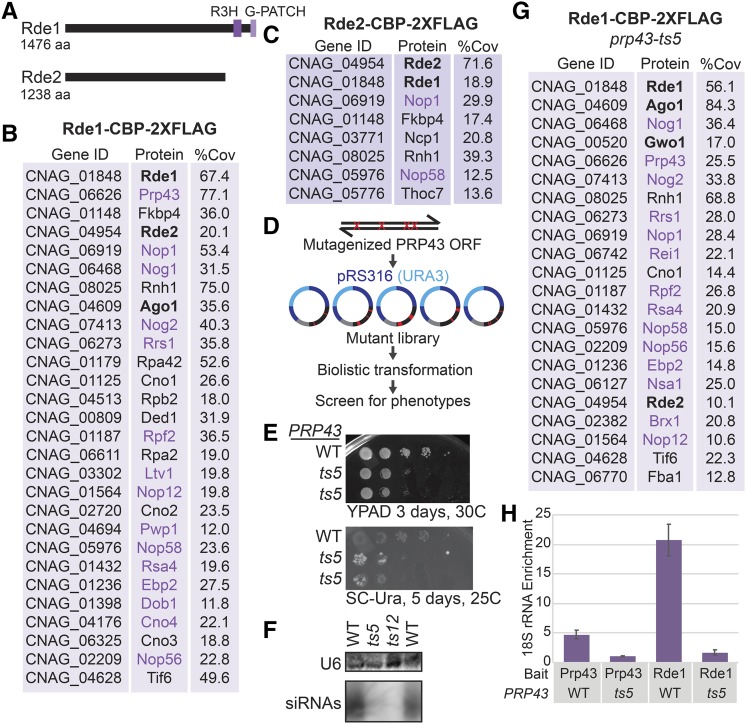
Figure 4.
A. Predicted domain structures of Rde1 and Rde2 determined by PSI-BLAST with an e-value of at most 1X10−5. B. Proteins detected in Rde1-CBP-2xFLAG purified material as determined by tandem IP-MS, named based on current C. neoformans annotation (FungiDB) or S. cerevisiae homolog. Three factors had no clear homolog, so we refer to them as putative C. neoformans NucleOlar protein 1-3 (Cno1, Cno2 and Cno3). Proteins in bold were identified in the screen. Proteins in purple are predicted nucleolar proteins, typically involved in rRNA processing and ribosome biogenesis. Percent coverage is the average between two replicates. Common contaminants and proteins with less than 10% coverage are excluded and proteins are in order of ascending total spectral counts. C. Proteins detected in Rde2-CBP-2xFLAG purified material (see B). D. Mutagenic library strategy for PRP43. E. Growth phenotype of wild type and prp43-ts5 C. neoformans strains bearing the ura5::HAR1 insertion on rich media and media lacking uracil. F. Loss of siRNAs against CNAG_06705 as determined by Northern analysis (see Fig. 2A). G. RT-qPCR of 18S rRNA associated with Rde1 (native FLAG affinity purification). Data are from three technical replicates and two biological replicates.