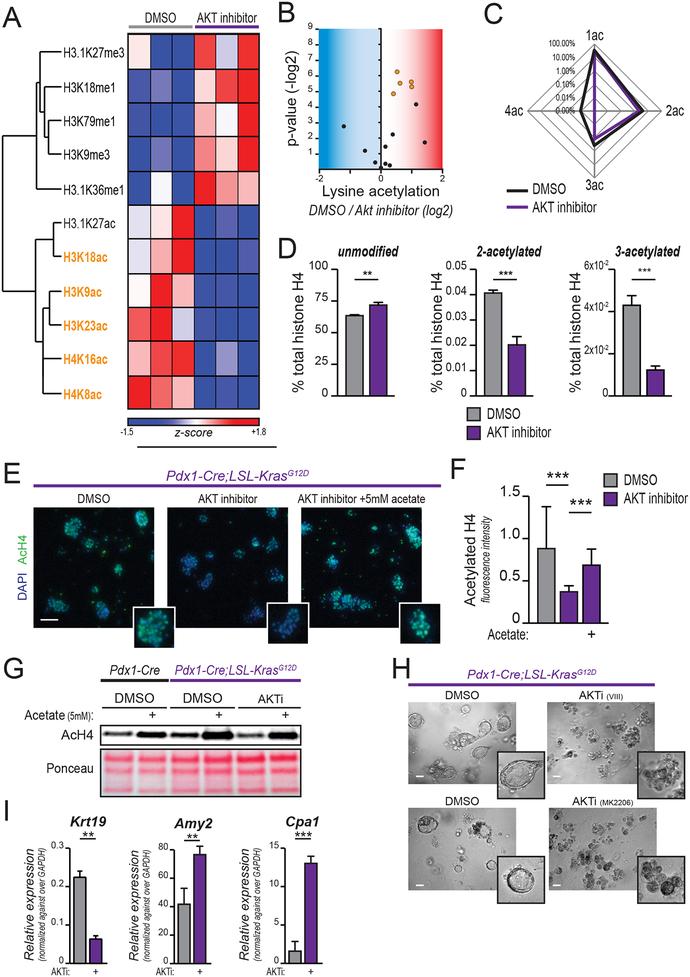
Figure 1: AKT inhibition reduces histone acetylation in KRASG12D-expressing cells.
A-D, Mass spectrometry-based profiling of histone modifications in primary murine PanIN-derived cells treated with AKT inhibitor (CAS-612847-09-3, 10 μM) for 24 hours. A, Heatmap shows histone marks most strongly regulated (see Supplementary Figure S1A for heat map of full dataset). Columns show biological replicates (n=3, each treatment). Histone acetyl marks highlighted in orange are those reaching p<0.05 (H3.1K27ac p=0.057). Complete histone acetylome represented in B, volcano plot. Each dot represents an acetylated residue. Blue area contains downregulated marks with AKTi, red area represents upregulated marks. Orange dots represent those reaching p<0.05, as in part A. C, Spider graph shows percentage of 1-, 2-, 3-, 4-acetylated histone H4 (lysine residues 5, 8, 12, 16) over total histone H4. D, bar graphs depict abundance of indicated poly-acetylated H4 peptides, along with unmodified histone H4. E, AcH4 staining of acinar cells isolated from 6–8-week-old KC mice (n=3), embedded in Matrigel, and treated as indicated for 24 hours. Scale bar, 50 μm. F, quantification of E (25 optical fields acquired per experimental replicate). G, AcH4 western blot of acinar cells treated 24 hours in indicated conditions (AKT selective inhibitor VIII, 10 μM; acetate, 5 mM). H, morphology (scale bar, 50 μm) of Matrigel-embedded acinar cells after 48 hours culture in the presence or absence of indicated AKT inhibitors (VIII, upper panel; MK2206, lower panel). I, mRNA expression of indicated genes quantified by qPCR for cells in H. Bar graphs depict mean, +/− SD of triplicates (*, p<0.05; **, p<0.01; ***, p<0.001).