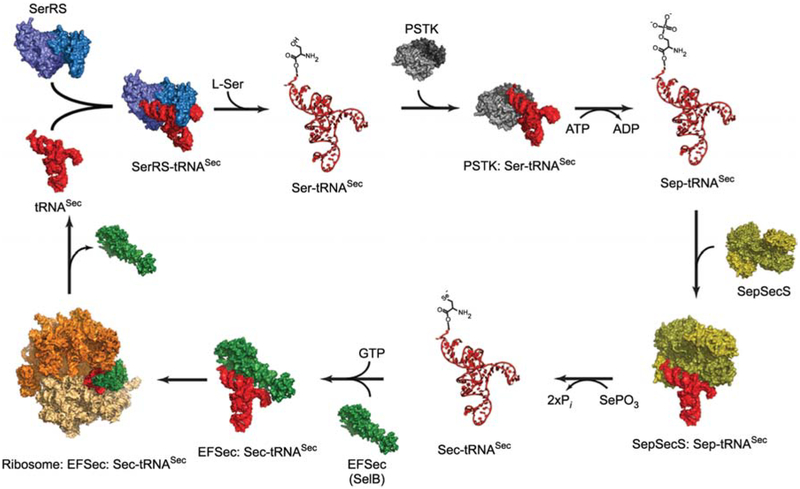
Figure 1.
A schematic diagram of the synthetic cycle of selenocysteine in eukaryotes.
The process begins with serylation of tRNASec (red) by SerRS (light and dark blue). PSTK (light and dark grey) then phosphorylates Ser-tRNASec and releases Sep-tRNASec and ADP. A SepSecS tetramer (gold and olive) subsequently binds Sep-tRNASec and catalyzes a two-step transformation of Sep into Sec using selenophosphate as the selenium donor. The final product, Sec-tRNASec, is delivered to the 80S ribosome (orange and beige) by the specialized elongation factor EFSec (green). Once the Sec residue is inserted into the nascent polypeptide chain, free tRNASec is released for another round of Sec synthesis. All molecules are shown in surface representation, whereas Ser-tRNASec, Sep-tRNASec and Sec-tRNASec are shown as ribbon diagrams. Crystal structures of the bacterial SerRS (Biou et al., 1994), the archaeal PSTK (Araiso et al., 2009), the human SepSecS-tRNASec complex (Palioura et al., 2009), the human tRNASec (Itoh et al., 2009), the archaeal SelB (Leibundgut et al., 2005) and that of the bacterial 70S ribosome in complex with EF-Tu (Schmeing et al., 2009) were used for modeling. Except for the SepSecS-tRNASec complex, all other complexes are proposed models and not true structures.