Figure 6. Gene expression is regulated by spatial positioning at the nuclear scale.
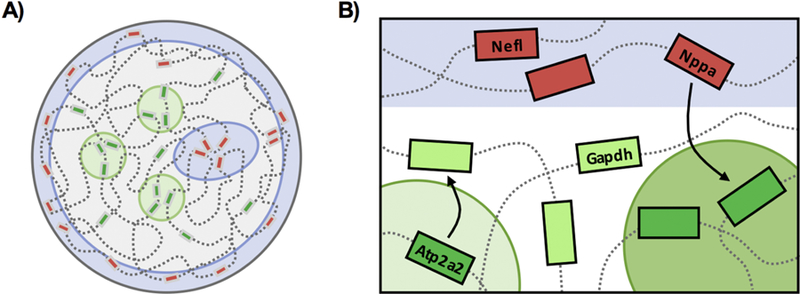
A) The nucleus is compartmentalized into active and inactive compartments. Inactive regions (red boxes) are found in heterochromatic regions (blue), which are heavily enriched at the nuclear lamina and centrally at chromocenters while highly transcribed regions (green boxes) are found at transcription factories (green). The cardiac nucleus contains stable RNA polymerase II transcription compartments enriched with active genes while genes outside of these sites in the nucleoplasm are expressed at low levels or poised for activation. Upon pathological stress, strength of transcription factory activity increases, without significant changes to number, through the recruitment of elongating RNA polymerase II molecules. B) The differential changes in gene expression are mediated by changes in gene positioning with respect to active (transcription factories, green) and inactive (heterochromatin, blue) compartments. Increases in gene expression correlate with the concentration of upregulated genes and RNA polymerase II molecules at discrete transcription sites.
