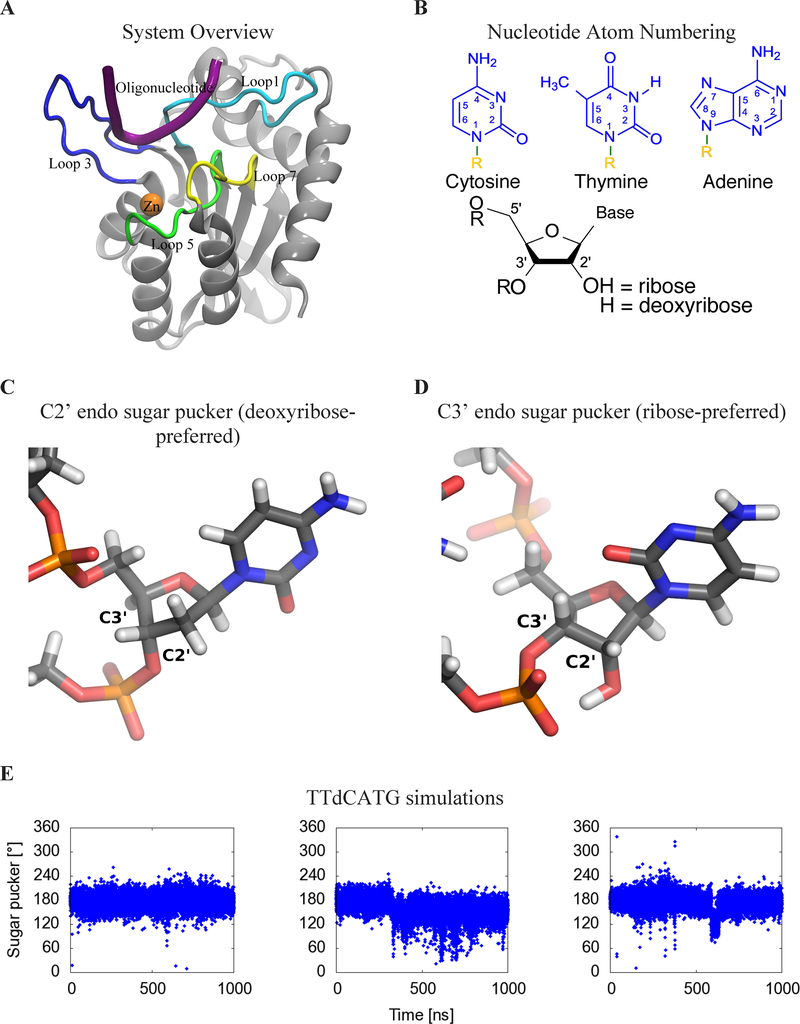
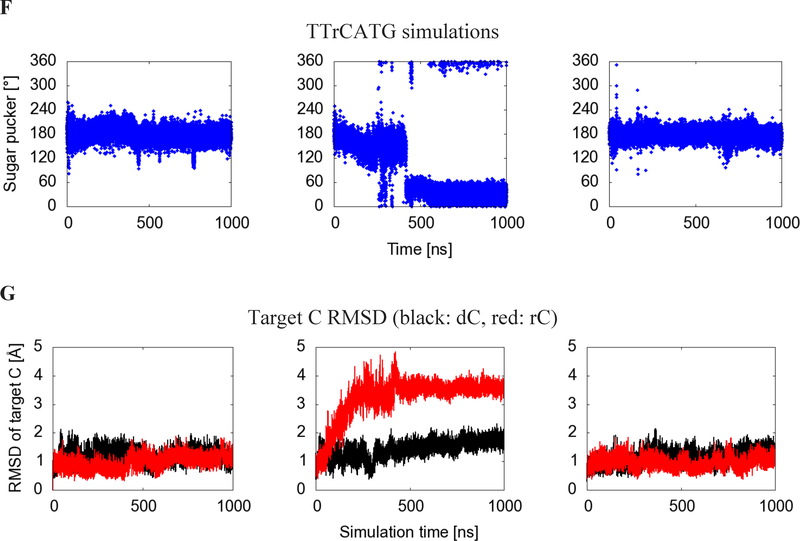
Figure 1:
(A) Cartoon view of the protein, labeling features of interest including loops 1 (light blue), 3 (blue), 5 (green), and 7 (yellow), the substrate oligonucleotide (purple), and the catalytic zinc (orange). (B) Numbering system used to identify atoms in nucleotides. R=DNA (C) Geometry of C2’ endo sugar pucker taken from the starting configuration of the dC simulation, and (D) C3’ endo sugar pucker taken from the second rC simulation. (E) Sugar pucker of the target C, measured in both A3B-dC and (F) A3B-rC simulations. In rC simulation replicate 2, the RNA transitions from a C2’ endo (DNA-preferred) to a C3’ endo (RNA-preferred) sugar pucker. (G) RMSD of target C compared to its initial pose in A3Bctd-DNA (black) and A3Bctd- DNA-rC (red) simulations.