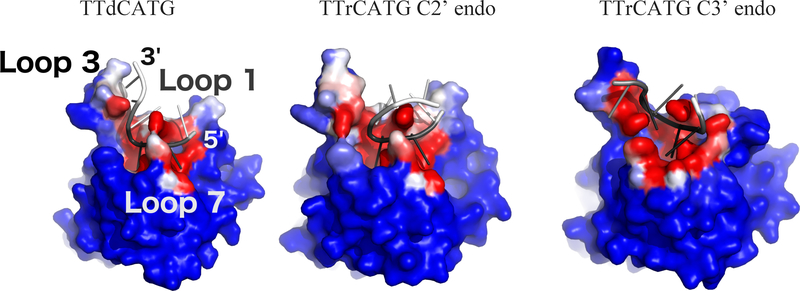
Figure 4:
Oligonucleotide interaction surface differences between DNA (left) and DNA-rC (center and right) simulations. Structures from the end of each simulation are shown. Each protein atom is colored by its frequency of contact (<5 Angstrom distance) with the oligonucleotide, on a color scale from blue (no contact) to white (50% contact) to red (100% contact). The oligonucleotides are colored by RMSF, with black corresponding to 0 angstroms and white to 5 Angstroms. The C3’ endo portion of the DNA-rC simulation shows a shift in position of the 0 and −1 nucleotides of the substrate, but low RMSD after the shift. Structures are RMSD-aligned.