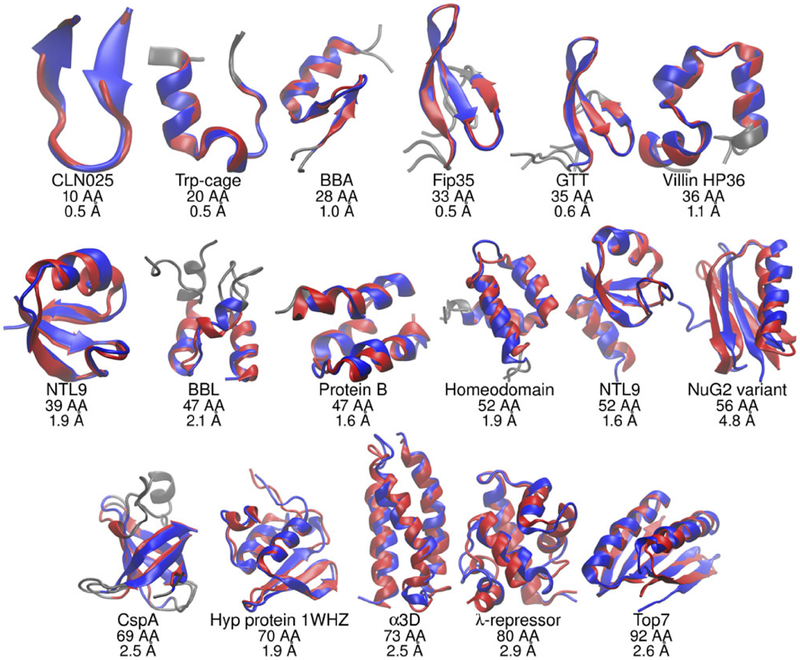
Figure 4.
GB-based molecular dynamics simulations (GB-neck2(81), Amber) of a number of proteins starting from completely unfolded states sample conformations (blue) that are close to the experimental native structures (red). Lowest RMSD distance to the experimental native structure, in Å, is indicated under each protein. Adapted from Ref.(79), courtesy of Carlos Simmerling.