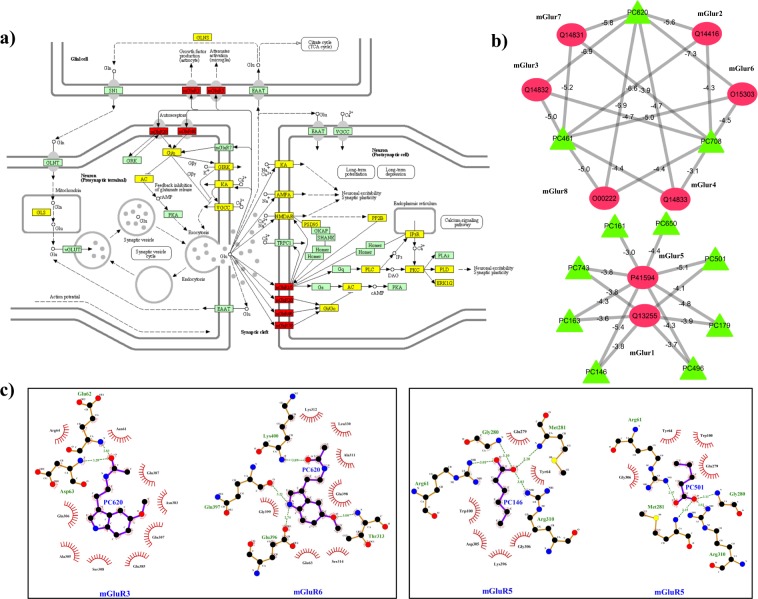
Figure 5.
(a) Glutamatergic synapse (path:hsa04724) pathway mapping of the protein targets of DPCs: The location of protein targets of DPCs in the KEGG pathway hsa04724 is represented in yellow and red coloured boxes, red ones are specific to mGluRs. Reprinted with permission from Kyoto Encyclopedia of Genes and Genome, https://www.kegg.jp/kegg/kegg1.html. (b) Sub-network of mGluRs and their regulatory DPCs: Green coloured nodes of the network correspond to 11 DPCs (PC146, PC161, PC163, PC179, PC461, PC496, PC501, PC620, PC650, PC708 and PC743) that possess the tendency to regulate 8 classes of mGluRs (Red coloured circular nodes). The binding energy values (kcal/mol) of the mGluRs and their regulatory DPCs are represented along with their corresponding edges in the network. For docking studies, the structure of mGluR6, mGluR4 and mGluR8 were modelled using PHYRE2 while for others they were obtained from RCSB-PDB with following PDB-IDs: 3KS9 (mGluR1), 4XAS (mGluR2), 6B7H (mGluR3), 3LMK (mGluR5) and 3MQ4 (mGluR7). (c) The interaction analysis of the mGluRs and their regulatory DPCs: Docked complexes with the best binding energy values are analysed for the hydrophobic interactions as well as hydrogen bonded residues using Ligplot+. The ligplots enclosed in boxes are the representative cases of multi-targeting and synergistic actions of selected phytochemicals respectively. Protein residues represented along the arcs are involved in the hydrophobic interactions whereas the residues involved in the hydrogen bonding are represented using dashed lines. The PC620 is shown to possess much negative binding energy compared to the other 10 DPCs and the interaction analysis highlights the importance of the acetamide group (-NHCOCH3) in the interaction.