Figure 3.
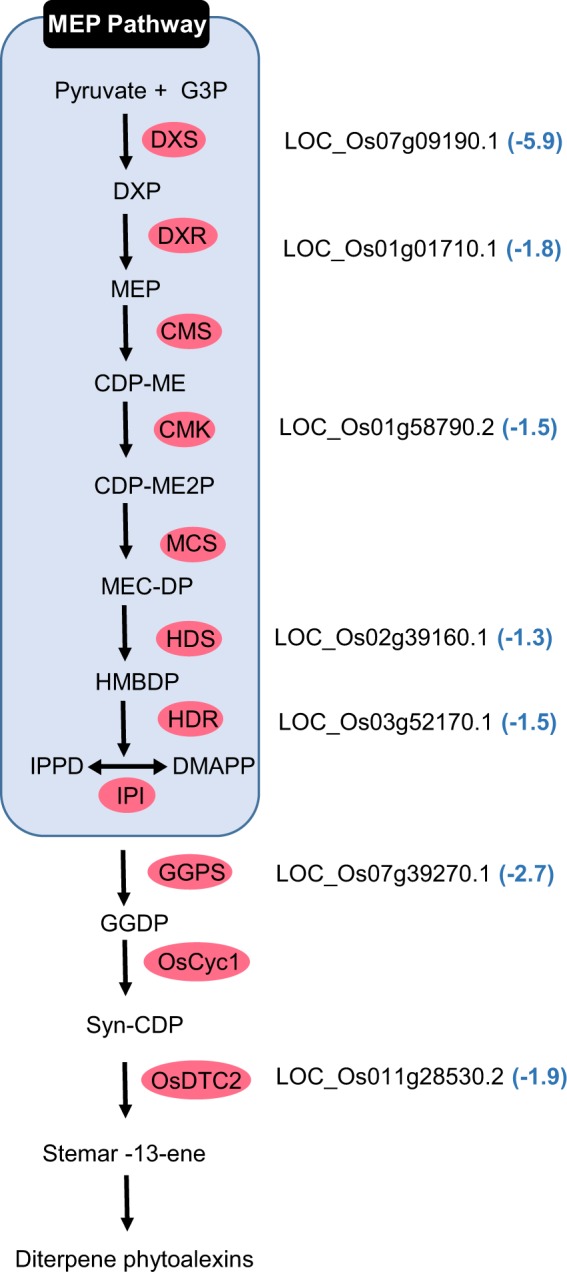
Isoprenoid synthesis genes down-regulated in rice roots colonised by H. seropedicae. The names of the genes differentially expressed are shown and the numbers in parentheses represent the fold change. The components of the MEP pathway leading to geranylgeranyl diphosphate synthesis and the diterpenoid- phytoalexin pathway are: G3P, glyceraldehyde-3-phosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5-diphospho)-2-C-methyl-D-erythritol; CDP-ME2P, 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; MEC-DP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBDP, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGDP, geranylgeranyl diphosphate; and CDP, copalyl diphosphate. Enzymes are indicated in rose coloured circles: DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, DXP reductoisomerase; CMS, CDP-ME synthase; CMK, CDP-ME kinase; MCS, MECDP synthase; HDS, HMBDP synthase; HDR, HMBDP reductase; IPI, IPP isomerase; GGPS, GGDP synthase; OsCyc1, syn-CDP synthase; OsCyc2, ent-CDP synthase; OsDTC2, stemar-13-ene synthase.
