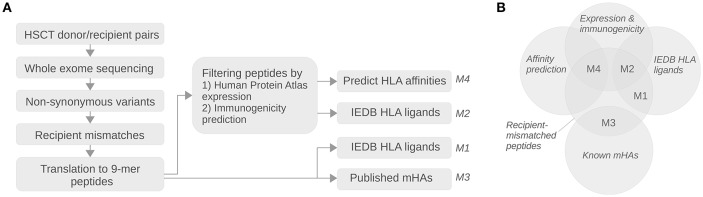
Figure 1.
Schematic diagram of the analysis pipeline. (A) The vertical arrows in top-down direction in the diagram show the processing steps for translating whole exome sequencing data into recipient-mismatched peptide sets. The horizontal arrows in left-to-right direction show the analysis steps involved in sub-setting the mismatched peptides into sets relevant for alloreactivity; in the first step the peptides are filtered for epithelial expression and immunogenicity, and in the second step they are intersected with experimental HLA ligand databases or HLA affinity predictions according to each pair's HLA class I type. Finally, the obtained HLA-presented peptide count estimates from the four analysis approaches (labeled as M1–M4; Table 2) are examined for possible association with chronic GvHD. (B) Euler diagram showing the analysis methods M1–M4 as intersections between peptide sets.