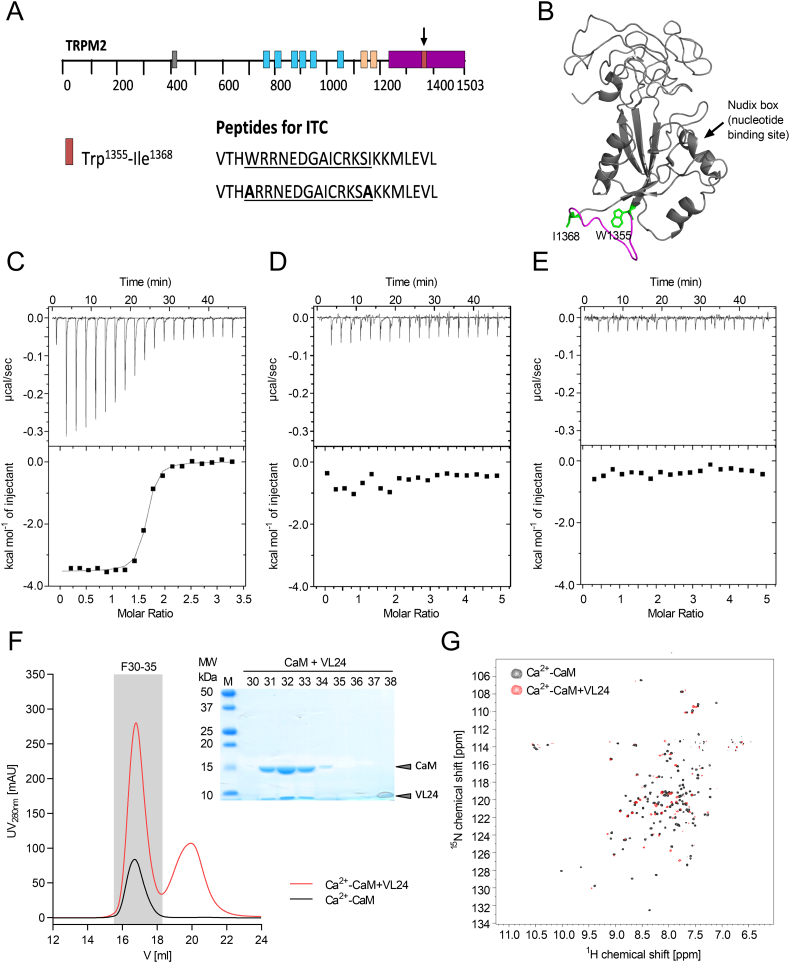
Fig. 1.
Identification of a new CaM-binding motif in the TRPM2 NudT9H domain. (A) Domain organization of the TRPM2 channel indicating the location of functional motifs in the amino acid sequence (grey box: known IQ-like motif, blue boxes: transmembrane helices, sand box: coiled coil domains, purple box: NudT9H domain, orange box: new CaM binding motif). The putative new CaM binding motif is marked by an arrow. The peptides used for ITC, NMR and fluorescence measurements are shown below. Peptide VTHWRRNEDGAICRKSIKKMLEVL is denoted VL24 throughout this study. (B) Structure of human NudT9H domain (from full-length hsTRPM2 cryo-EM structure (pdb:6MIX)). The putative CaM binding motif (magenta loop with anchor residues in green) is located on the surface of the NudT9H domain. The location of the Nudix box, the putative ADPR binding site is indicated by an arrow. (C–E) ITC measurements of CaM and the respective peptide. (C) Binding of CaM to peptide VL24 in the presence of 2 mM Ca2+ revealed a Kd of 110 nM (±18 nM). (D) VL24 with mutated anchor residues does not bind to Ca2+-CaM. (E) VL24 does not bind to CaM in the absence of Ca2+. (F) Purification of the complex of Ca2+-CaM and VL24 by SEC. Chromatography profiles of CaM alone and in complex with the peptide (left) and the SDS-PAGE of the corresponding complex fractions (right). (G) BEST-TROSY NMR spectrum of 15N-Ca2+-CaM alone (black) and in complex with access unlabelled VL24 peptide (red). Large perturbation of chemical shifts or disappearance of many peaks in the complex indicates significant conformational changes upon binding.